4UIS
 
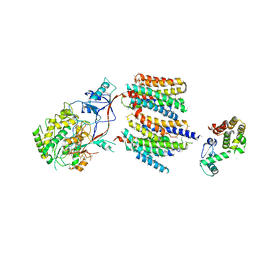 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XOI
 
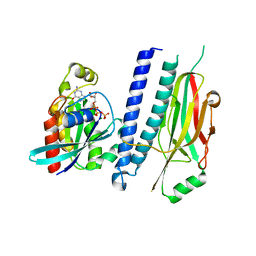 | | Structure of hsAnillin bound with RhoA(Q63L) at 2.1 Angstroms resolution | | Descriptor: | Actin-binding protein anillin, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sun, L, Guan, R, Lee, I.-J, Liu, Y, Chen, M, Wang, J, Wu, J, Chen, Z. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
9JDR
 
 | |
9M2H
 
 | |
5KNN
 
 | |
7SA0
 
 | | Crystal structure of CDK2 liganded with compound EF4195 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(3R)-2,3,4,9-tetrahydro-1H-carbazol-3-yl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-21 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of selective allosteric inhibitors of cyclin-dependent kinase 2 (CDK2)
To Be Published
|
|
7S9X
 
 | | Crystal structure of CDK2 liganded with compound WN378 | | Descriptor: | 2-[(9H-carbazol-3-yl)amino]-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-21 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of selective allosteric inhibitors of cyclin-dependent kinase 2 (CDK2)
To Be Published
|
|
5T76
 
 | | A fragment of a human tRNA synthetase | | Descriptor: | Alanine--tRNA ligase, cytoplasmic | | Authors: | Sun, L, Schimmel, P. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5T5S
 
 | | A fragment of a human tRNA synthetase | | Descriptor: | Alanine--tRNA ligase, cytoplasmic | | Authors: | Sun, L, Schimmel, P. | | Deposit date: | 2016-08-31 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3JA7
 
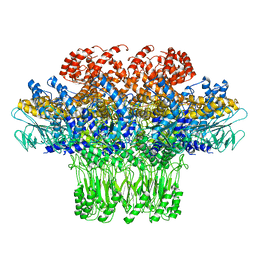 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
9J14
 
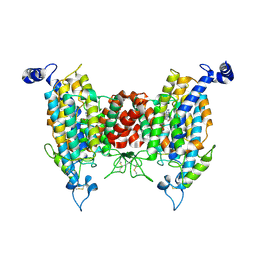 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state at pH 5.5 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state at pH 5.5
To Be Published
|
|
9J18
 
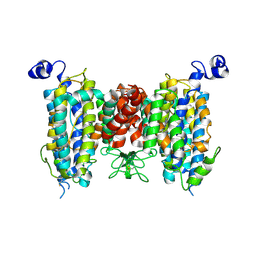 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-2 at pH 7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-2 at pH 7.4
To Be Published
|
|
9J13
 
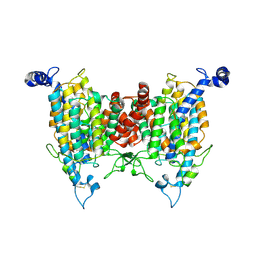 | |
9J16
 
 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 7.4 | | Descriptor: | ADENINE, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 7.4
To Be Published
|
|
9J17
 
 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-1 at pH 7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-1 at pH 7.4
To Be Published
|
|
9J15
 
 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 5.5 | | Descriptor: | ADENINE, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 5.5
To Be Published
|
|
9J12
 
 | |
6DIZ
 
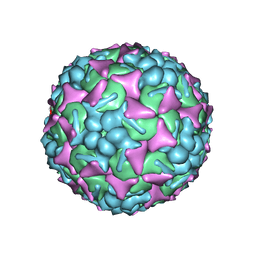 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | Deposit date: | 2018-05-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
1P5J
 
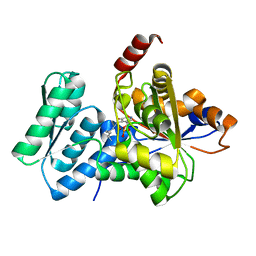 | | Crystal Structure Analysis of Human Serine Dehydratase | | Descriptor: | L-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sun, L, Liu, Y, Rao, Z. | | Deposit date: | 2003-04-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of human serine dehydratase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2LJ4
 
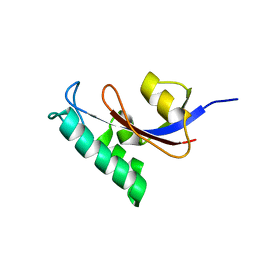 | | Solution structure of the TbPIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase/rotamase, putative | | Authors: | Sun, L, Lin, D, Zhao, Y. | | Deposit date: | 2011-09-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural analysis of the single-domain parvulin TbPin1.
Plos One, 7, 2012
|
|
3V0U
 
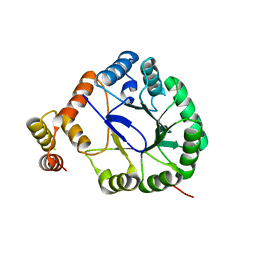 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3V0S
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UA4
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 5 | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
4JPP
 
 | | Bacteriophage phiX174 H protein residues 143-282 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
