8RG2
 
 | |
8RFZ
 
 | |
8RII
 
 | |
1YOP
 
 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
6WXV
 
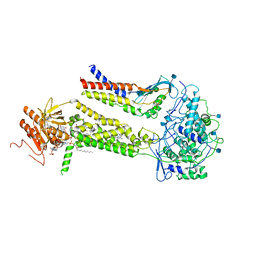 | | CryoEM structure of mouse DUOX1-DUOXA1 complex in the presence of NADPH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sun, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of mouse DUOX1-DUOXA1 provide mechanistic insights into enzyme activation and regulation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WXU
 
 | | CryoEM structure of mouse DUOX1-DUOXA1 complex in the dimer-of-dimer state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sun, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of mouse DUOX1-DUOXA1 provide mechanistic insights into enzyme activation and regulation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WXR
 
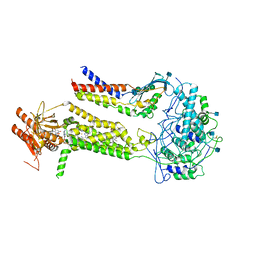 | | CryoEM structure of mouse DUOX1-DUOXA1 complex in the absence of NADPH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, Dual oxidase maturation factor 1, ... | | Authors: | Sun, J. | | Deposit date: | 2020-05-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of mouse DUOX1-DUOXA1 provide mechanistic insights into enzyme activation and regulation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3OIQ
 
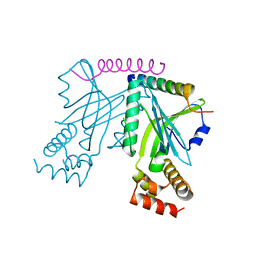 | | Crystal structure of yeast telomere protein Cdc13 OB1 and the catalytic subunit of DNA polymerase alpha Pol1 | | Descriptor: | Cell division control protein 13, DNA polymerase alpha catalytic subunit A | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
3OIP
 
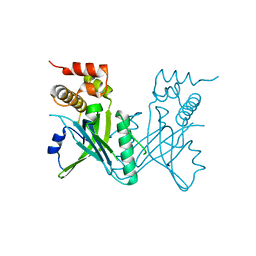 | | Crystal structure of Yeast telomere protein Cdc13 OB1 | | Descriptor: | Cell division control protein 13 | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
3KF6
 
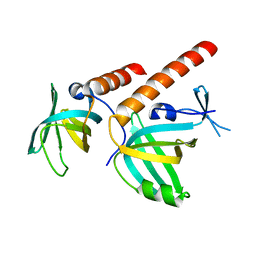 | | Crystal structure of S. pombe Stn1-ten1 complex | | Descriptor: | Protein stn1, Protein ten1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
3KEY
 
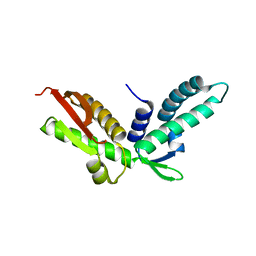 | | Crystal structure of S. cerevisiae Stn1 C-terminal | | Descriptor: | Protein STN1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
3KF8
 
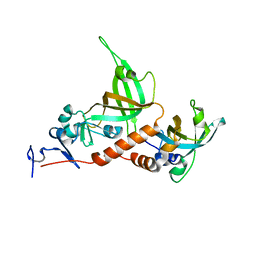 | | Crystal structure of C. tropicalis Stn1-ten1 complex | | Descriptor: | Protein stn1, Protein ten1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
8EYT
 
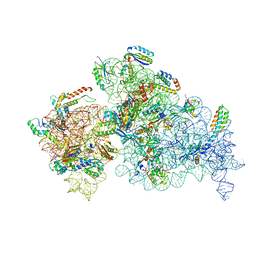 | | 30S_delta_ksgA+KsgA complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sun, J, Kinman, L.F, Jahagirdar, D, Ortega, J, Davis, J.H. | | Deposit date: | 2022-10-28 | | Release date: | 2023-09-06 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8JEP
 
 | | Crystal structure of Ociperlimab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEL
 
 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form I | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEO
 
 | | Crystal structure of TIGIT in complexed with Tiragolumab | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEN
 
 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form II | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEQ
 
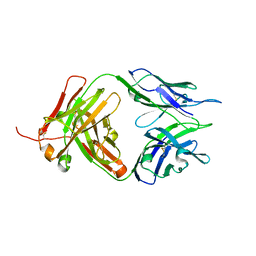 | | Crystal structure of Tiragolumab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
6CW2
 
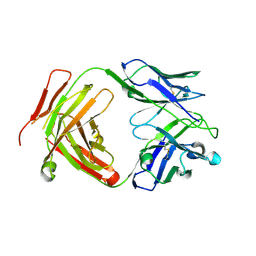 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4OH3
 
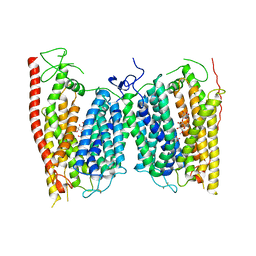 | | Crystal structure of a nitrate transporter | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, NITRATE ION, Nitrate transporter 1.1 | | Authors: | Sun, J, Bankston, J.R, Payandeh, J, Hinds, T.R, Zagotta, W.N, Zheng, N. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the plant dual-affinity nitrate transporter NRT1.1.
Nature, 507, 2014
|
|
6CW3
 
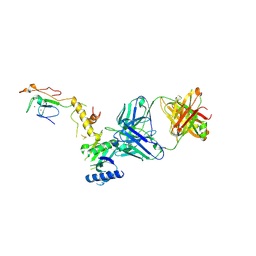 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8XHE
 
 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8XG1
 
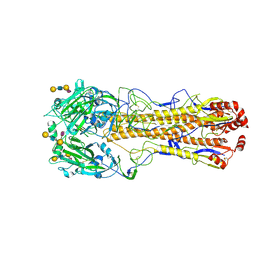 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8XHG
 
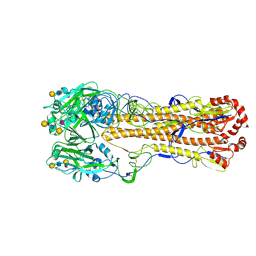 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8XHH
 
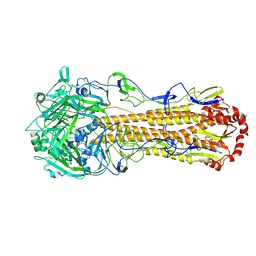 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
