8JLO
 
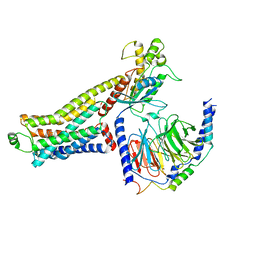 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLJ
 
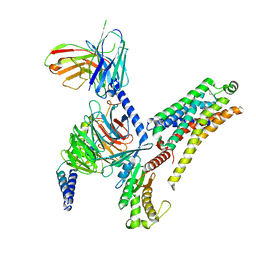 | | T1AM-bound mTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSO
 
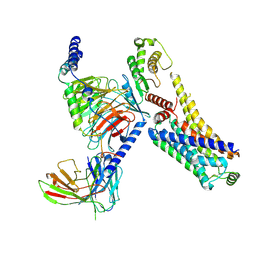 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLR
 
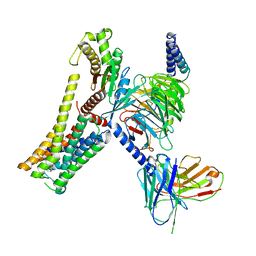 | | A77636-bound hTAAR1-Gs protein complex | | Descriptor: | (1~{S},3~{R})-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1~{H}-isochromene-5,6-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSP
 
 | | Ulotaront(SEP-363856)-bound Serotonin 1A (5-HT1A) receptor-Gi complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, 5-hydroxytryptamine receptor 1A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6E3K
 
 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6E3L
 
 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6CT7
 
 | |
3LKJ
 
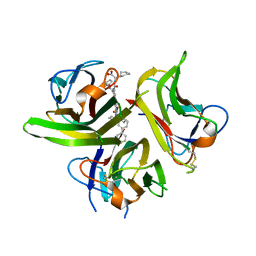 | | Small Molecule Inhibition of the TNF Family Cyokine CD40 Ligand Through a Subunit Fracture Mechanism | | Descriptor: | (2R)-{[(2'-[(biphenyl-3-ylmethyl)carbamoyl]-6'-{[(2R)-2-(pyrrolidin-1-ylmethyl)pyrrolidin-1-yl]carbonyl}-6-{[(2R)-2-(1H-pyrrol-1-ylmethyl)pyrrolidin-1-yl]carbonyl}-4,4'-bipyridin-2-yl)carbonyl]amino}(cyclohexyl)ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand | | Authors: | Silvian, L.F, Whitty, A. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small Molecule Inhibition of the TNF Family Cytokine CD40 Ligand through a Subunit Fracture Mechanism.
Acs Chem.Biol., 6, 2011
|
|
6WDP
 
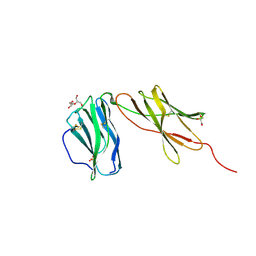 | | Interleukin 12 receptor subunit beta-1 | | Descriptor: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | Authors: | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WDQ
 
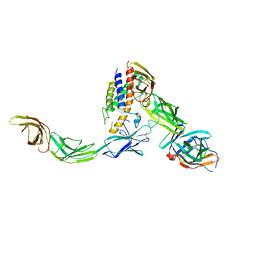 | | IL23/IL23R/IL12Rb1 signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | Authors: | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WEO
 
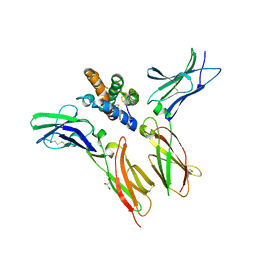 | | IL-22 Signaling Complex with IL-22R1 and IL-10Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saxton, R.A, Jude, K.M, Henneberg, L.T, Garcia, K.C. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design.
Immunity, 54, 2021
|
|
7RAA
 
 | | Designed StabIL-2 seq15 | | Descriptor: | Interleukin-2, MAGNESIUM ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RA9
 
 | | Designed StabIL-2 seq1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Interleukin-2, PHOSPHATE ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YWA
 
 | | Structure of DinI in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA damage-inducible protein I, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
4G3Y
 
 | | Crystal structure of TNF-alpha in complex with Infliximab Fab fragment | | Descriptor: | Tumor necrosis factor, infliximab Fab H, infliximab Fab L | | Authors: | Liang, S.Y, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-07-15 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
J.Biol.Chem., 288, 2013
|
|
8GMU
 
 | | Structure of lambda repressor in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMT
 
 | | Structure of UmuD in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA polymerase V subunit UmuD, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMS
 
 | | Structure of LexA in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), LexA repressor, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WQN
 
 | |
5WQO
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
