1ACY
 
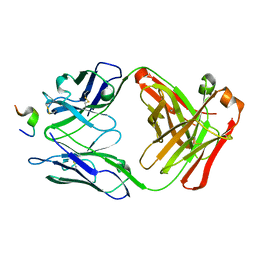 | |
1GGI
 
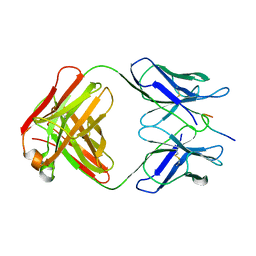 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1VAD
 
 | |
2CKK
 
 | | High resolution crystal structure of the human kin17 C-terminal domain containing a kow motif | | Descriptor: | ACETATE ION, IODIDE ION, KIN17 | | Authors: | le Maire, A, Schiltz, M, Pinon-Lataillade, G, Stura, E, Couprie, J, Gondry, M, Angulo-Mora, J, Zinn-Justin, S. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Tandem of SH3-Like Domains Participates in RNA Binding in Kin17, a Human Protein Activated in Response to Genotoxics.
J.Mol.Biol., 364, 2006
|
|
3GAR
 
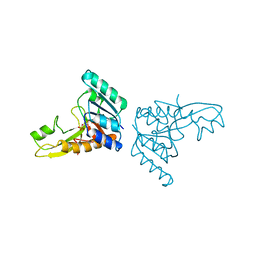 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
7FD1
 
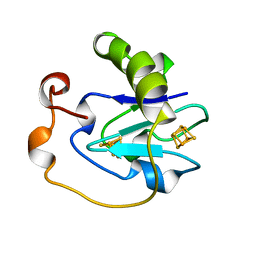 | | 7-FE FERREDOXIN FROM AZOTOBACTER VINELANDII AT PH 8.5, 100 K, 1.35 A | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (7-FE FERREDOXIN I) | | Authors: | Stout, C.D, Stura, E.A, Mcree, D.E. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidized and reduced Azotobacter vinelandii ferredoxin I at 1.4 A resolution: conformational change of surface residues without significant change in the [3Fe-4S]+/0 cluster.
Biochemistry, 38, 1999
|
|
7ANQ
 
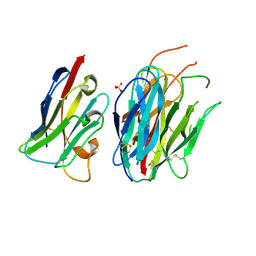 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
5M05
 
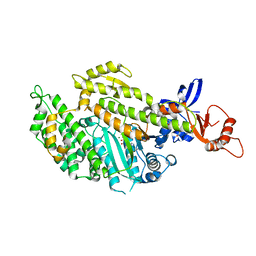 | | Chicken smooth muscle myosin motor domain co-crystallized with the specific CK-571 inhibitor, MgADP form | | Descriptor: | 4-{[(2-chloro-3-fluorobenzyl)carbamoyl](methyl)amino}-3,4-dideoxy-5-O-(isoquinolin-3-ylcarbamoyl)-D-erythro-pentitol, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sirigu, S, Hartman, J, Houdusse, A. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | Highly selective inhibition of myosin motors provides the basis of potential therapeutic application.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1FRG
 
 | |
1GAR
 
 | | TOWARDS STRUCTURE-BASED DRUG DESIGN: CRYSTAL STRUCTURE OF A MULTISUBSTRATE ADDUCT COMPLEX OF GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE AT 1.96 ANGSTROMS RESOLUTION | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, N-[4-[[3-(2,4-DIAMINO-1,6-DIHYDRO-6-OXO-4-PYRIMIDINYL)-PROPYL]-[2-((2-OXO-2-((4-PHOSPHORIBOXY)-BUTYL)-AMINO)-ETHYL)-THIO-ACETYL]-AMINO]BENZOYL]-1-GLUTAMIC ACID | | Authors: | Wilson, I.A, Klein, C, Chen, P, Arevalo, J.H. | | Deposit date: | 1994-12-08 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 A resolution.
J.Mol.Biol., 249, 1995
|
|
1IGF
 
 | |
1F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
3MKO
 
 | | Crystal Structure of the Lymphocytic Choriomeningitis Virus Membrane Fusion Glycoprotein GP2 in its Postfusion Conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Glycoprotein C, ... | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2010-04-15 | | Release date: | 2011-04-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1A3L
 
 | |
15C8
 
 | | CATALYTIC ANTIBODY 5C8, FREE FAB | | Descriptor: | IGG 5C8 FAB (HEAVY CHAIN), IGG 5C8 FAB (LIGHT CHAIN) | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Conformational Changes in a Catalytic Antibody: Comparison of the Bound and Unbound Structure of Fab 5C8
To be Published
|
|
7OVY
 
 | | Crystal structure of a dimeric based inhibitor JG34 in complex with the MMP-12 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Ciccone, L, Rossello, A, Vera, L, Nuti, E, Stura, E.A. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of Dimeric Arylsulfonamides as Potent ADAM8 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1AXT
 
 | |
1CD1
 
 | |
2F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120) (MN ISOLATE) | | Descriptor: | 1-IMINO-5-PENTANONE, PROTEIN (HIV-1 GP120), PROTEIN (IGG1 FAB 58.2 ANTIBODY (HEAVY CHAIN)), ... | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-23 | | Release date: | 1999-02-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
2H5F
 
 | | Denmotoxin: A the three-finger toxin from colubrid snake Boiga dendrophila with bird-specific activity | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Pawlak, J, Kini, R.M, Stura, E.A. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity.
J.Biol.Chem., 281, 2006
|
|
6S8C
 
 | |
1URZ
 
 | |
2GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
1FPT
 
 | |
3MK1
 
 | | Refinement of placental alkaline phosphatase complexed with nitrophenyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
