2OAU
 
 | | Mechanosensitive Channel of Small Conductance (MscS) | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Rees, D.C, Bass, R.B, Steinbacher, S, Strop, P, Barclay, M.T. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
CURRENT TOPICS IN MEMBRANES, 58, 2007
|
|
6GK7
 
 | | Crystal structure of anti-tau antibody dmCBTAU-27.1, double mutant (S31Y, T100I) of CBTAU-27.1, in complex with Tau peptide A8119B (residues 299-318) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-27.1(S31Y,T100I), HUMAN TAU PEPTIDE A8119 RESIDUES 299-318 | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
6E5B
 
 | | Human Immunoproteasome 20S particle in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Steinbacher, S, Augustin, M, Blaesse, M, Harris, S.F. | | Deposit date: | 2018-07-19 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Design and Evaluation of Highly Selective Human Immunoproteasome Inhibitors Reveal a Compensatory Process That Preserves Immune Cell Viability.
J.Med.Chem., 62, 2019
|
|
6GK8
 
 | | Crystal structure of anti-tau antibody dmCBTAU-28.1, double mutant (S32R, E35K) of CBTAU-28.1, in complex with Tau peptide A7731 (residues 52-71) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-28.1(S32R;E35K), TAU PEPTIDE A7731 (RESIDUES 52-71) | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
5IWG
 
 | | HDAC2 WITH LIGAND BRD4884 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5IX0
 
 | | HDAC2 WITH LIGAND BRD7232 | | Descriptor: | (3-exo)-N-(4-amino-4'-fluoro[1,1'-biphenyl]-3-yl)-8-oxabicyclo[3.2.1]octane-3-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
1CLW
 
 | | TAILSPIKE PROTEIN FROM PHAGE P22, V331A MUTANT | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S, Baxa, U, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-05-04 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
3TG6
 
 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
1KZL
 
 | | Riboflavin Synthase from S.pombe bound to Carboxyethyllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, MERCURY (II) ION, Riboflavin Synthase | | Authors: | Gerhardt, S, Schott, A.K, Kairies, N, Cushman, M, Illarionov, B, Eisenreich, W, Bacher, A, Huber, R, Steinbacher, S, Fischer, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on the Reaction Mechanism of Riboflavin Synthase; X-Ray Crystal Structure of a Complex with 6-Carboxyethyl-7-Oxo-8-Ribityllumazine
STRUCTURE, 10, 2002
|
|
2JDW
 
 | |
4JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | ARGININE, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
4H58
 
 | | BRAF in complex with compound 3 | | Descriptor: | CHLORIDE ION, N-(4-{[(2-methoxyethyl)amino]methyl}phenyl)-6-(pyridin-4-yl)quinazolin-2-amine, Serine/threonine-protein kinase B-raf | | Authors: | Vasbinder, M, Aquila, B, Augustin, M, Chueng, T, Cook, D, Drew, L, Fauber, B, Glossop, S, Godin, R, Grondine, M, Hennessy, E, Johannes, J, Lee, S, Lyne, P, Moertl, M, Omer, C, Palakurthi, S, Pontz, T, Read, J, Sha, L, Shen, M, Steinbacher, S, Wang, H, Wu, A, Ye, M, Bagal, B. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf(V600E) Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3JDW
 
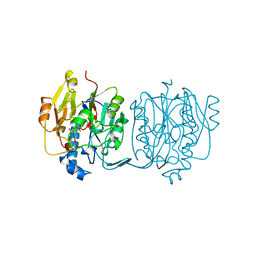 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
5DXH
 
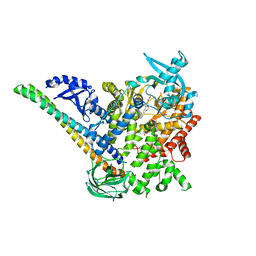 | | p110alpha/p85alpha with compound 5 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, methyl {2-[4-(2-chlorophenyl)-4H-1,2,4-triazol-3-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepin-8-yl}carbamate | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
4NMH
 
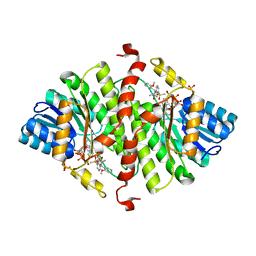 | | 11-beta-HSD1 in complex with a 3,3-Di-methyl-azetidin-2-one | | Descriptor: | (4S)-4-(2-methoxyphenyl)-3,3-dimethyl-1-[3-(methylsulfonyl)phenyl]azetidin-2-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McCoull, W, Augustin, M, Blake, C, Ertan, A, Kilgour, E.K, Krapp, S, Moore, J.E, Newcombe, N.J, Packer, M.J, Rees, A, Revill, J, Scott, J.S, Selmi, N, Gerhardt, S, Ogg, D.J, Steinbacher, S, Whittamore, P.R.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and optimisation of 3,3-dimethyl-azetidin-2-ones as potent and selective inhibitors of 11 beta-hydroxysteroid dehydrogenase type 1 (11-beta-HSD1)
TO BE PUBLISHED
|
|
5DXU
 
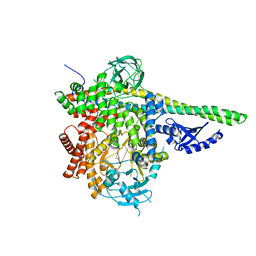 | | p110delta/p85alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
5DXT
 
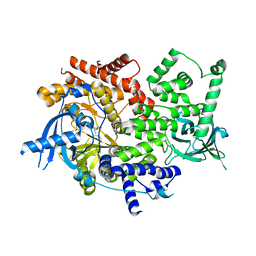 | | p110alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, 1,2-ETHANEDIOL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
3RO5
 
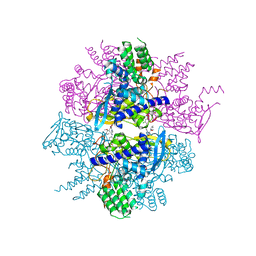 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5T8F
 
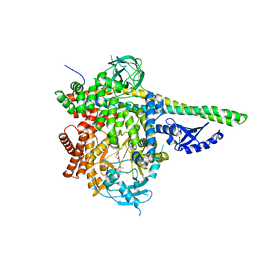 | | p110delta/p85alpha with taselisib (GDC-0032) | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Moertl, M, Steinbacher, S, Eigenbrot, C. | | Deposit date: | 2016-09-07 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
3LU8
 
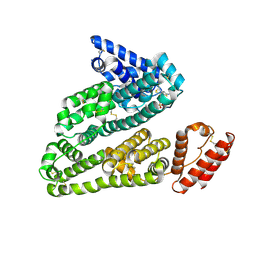 | | Human serum albumin in complex with compound 3 | | Descriptor: | N-[5-(5-{[(2,4-dimethyl-1,3-thiazol-5-yl)sulfonyl]amino}-6-fluoropyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]acetamide, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
3LU6
 
 | | Human serum albumin in complex with compound 1 | | Descriptor: | Serum albumin, [(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]acetic acid | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
3LU7
 
 | | Human serum albumin in complex with compound 2 | | Descriptor: | 4-[(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]butanoic acid, PHOSPHATE ION, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
6GL3
 
 | | Crystal structure of human Phosphatidylinositol 4-kinase III beta (PI4KIIIbeta) in complex with ligand 44 | | Descriptor: | (3~{S})-4-(6-azanyl-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl)-~{N}-(4-methoxy-2-methyl-phenyl)-3-methyl-piperazine-1-carboxamide, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Lammens, A, Augustin, M, Steinbacher, S, Reuberson, J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of a Potent, Orally Bioavailable PI4KIII beta Inhibitor (UCB9608) Able To Significantly Prolong Allogeneic Organ Engraftment in Vivo.
J. Med. Chem., 61, 2018
|
|
1JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | BETA-MERCAPTOETHANOL, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-22 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
6B8A
 
 | | Crystal structure of MvfR ligand binding domain in complex with M64 | | Descriptor: | 2-[(5-nitro-1H-benzimidazol-2-yl)sulfanyl]-N-(4-phenoxyphenyl)acetamide, COBALT HEXAMMINE(III), DNA-binding transcriptional regulator | | Authors: | Kitao, T, Steinbacher, S, Maskos, K, Blaesse, M, Rahme, L.G. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Insights into Function and Competitive Inhibition ofPseudomonas aeruginosaMultiple Virulence Factor Regulator.
MBio, 9, 2018
|
|
