6W3P
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with beta-methylnorleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3X
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-valine | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, SULFATE ION, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3T
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-norvaline | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, NORVALINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
4MDU
 
 | | Crystal structure of apo-Annexin (Sm)1 | | Descriptor: | Annexin | | Authors: | Hofmann, A. | | Deposit date: | 2013-08-23 | | Release date: | 2014-02-26 | | Last modified: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and immunological properties of the first annexin from Schistosoma mansoni: insights into the structural integrity of the schistosomal tegument.
Febs J., 281, 2014
|
|
4MDV
 
 | | Crystal structure of calcium-bound annexin (Sm)1 | | Descriptor: | Annexin, CALCIUM ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-08-23 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and immunological properties of the first annexin from Schistosoma mansoni: insights into the structural integrity of the schistosomal tegument.
Febs J., 281, 2014
|
|
5EMS
 
 | | Crystal Structure of an iodinated insulin analog | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Lawrence, M.C, Pandyarajan, V, Wan, Z, Weiss, M.A. | | Deposit date: | 2015-11-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extending Halogen-based Medicinal Chemistry to Proteins: IODO-INSULIN AS A CASE STUDY.
J. Biol. Chem., 291, 2016
|
|
4NIB
 
 | |
4OGA
 
 | |
4P65
 
 | | Crystal structure of an cyclohexylalanine substituted insulin analog. | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Pandyarajan, V, Wan, Z, Weiss, M.A. | | Deposit date: | 2014-03-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aromatic Anchor at an Invariant Hormone-Receptor Interface: FUNCTION OF INSULIN RESIDUE B24 WITH APPLICATION TO PROTEIN DESIGN.
J.Biol.Chem., 289, 2014
|
|
2MLI
 
 | |
6MTU
 
 | | Crystal structure of human Scribble PDZ1:pMCC complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Colorectal mutant cancer protein, ... | | Authors: | Caria, S, Stewart, B.Z, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural analysis of phosphorylation-associated interactions of human MCC with Scribble PDZ domains.
Febs J., 286, 2019
|
|
6MTV
 
 | | Crystal structure of human Scribble PDZ1:MCC complex | | Descriptor: | Colorectal mutant cancer protein, DI(HYDROXYETHYL)ETHER, Protein scribble homolog, ... | | Authors: | Caria, S, Stewart, B.Z, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural analysis of phosphorylation-associated interactions of human MCC with Scribble PDZ domains.
Febs J., 286, 2019
|
|
6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | Authors: | Gulbis, J.M, Clarke, O.B. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
1IEQ
 
 | | CRYSTAL STRUCTURE OF BARLEY BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, beta-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1IEV
 
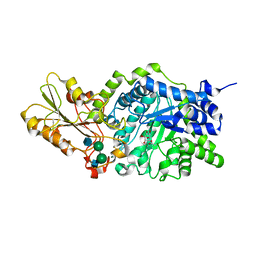 | | CRYSTAL STRUCTURE OF BARLEY BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1 IN COMPLEX WITH CYCLOHEXITOL | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
4BDU
 
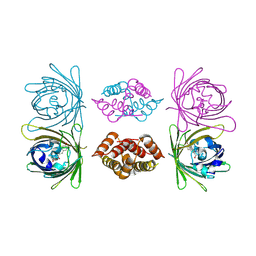 | | Bax BH3-in-Groove dimer (GFP) | | Descriptor: | GREEN FLUORESCENT PROTEIN, APOPTOSIS REGULATOR BAX | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-02-13 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD6
 
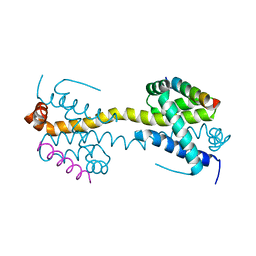 | | Bax domain swapped dimer in complex with BaxBH3 | | Descriptor: | APOPTOSIS REGULATOR BAX | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD8
 
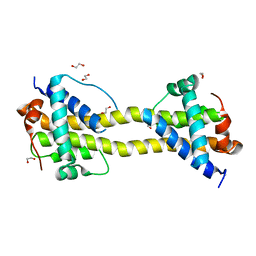 | | Bax domain swapped dimer induced by BimBH3 with CHAPS | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BAX, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD2
 
 | | Bax domain swapped dimer in complex with BidBH3 | | Descriptor: | APOPTOSIS REGULATOR BAX, BH3-INTERACTING DOMAIN DEATH AGONIST | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-04 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD7
 
 | | Bax domain swapped dimer induced by octylmaltoside | | Descriptor: | APOPTOSIS REGULATOR BAX, CHLORIDE ION, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4C52
 
 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
4C5D
 
 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (42) | | Descriptor: | (R)-3-(4-BROMOBENZYLTHIO)-2-(3-(3-((2,4-DIFLUOROPHENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
6BWK
 
 | |
2K9E
 
 | |
2MPI
 
 | | Solution structure of B24G insulin | | Descriptor: | insulin chain A, insulin chain B | | Authors: | Yang, Y, Wickramasinghe, N.P, Hua, Q, Weiss, M.A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protective hinge in insulin opens to enable its receptor engagement.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
