2LPA
 
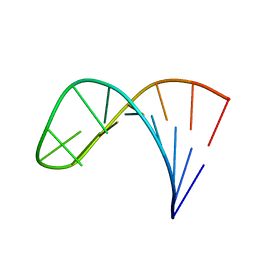 | | Mutant of the sub-genomic promoter from Brome Mosaic Virus | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*UP*CP*UP*UP*C)-3') | | Authors: | Skov, J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
2LP9
 
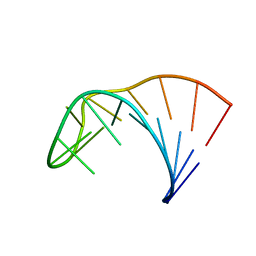 | | Pseudo-triloop from the sub-genomic promoter of Brome Mosaic Virus | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*AP*UP*CP*UP*UP*C)-3') | | Authors: | Skov, J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
6YLN
 
 | | mTurquoise2 SG P212121 - Directional optical properties of fluorescent proteins | | Descriptor: | EGFP, POTASSIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLO
 
 | | mTurquoise2 - Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | POTASSIUM ION, TETRAETHYLENE GLYCOL, mTurquoise2_C2221 | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLM
 
 | | mCherry | | Descriptor: | CHLORIDE ION, mCherry | | Authors: | Myskova, J, Rybakova, M, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLP
 
 | | EGFP_in_Acidic_env Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | PHOSPHATE ION, eGFP_in_Acidic_env | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLQ
 
 | | EGFP in neutral pH, Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | GLYCEROL, MAGNESIUM ION, eGFP | | Authors: | Myskova, J, Rybakova, M, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLS
 
 | | mEos4b - Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | CHLORIDE ION, Green to red photoconvertible GFP-like protein EosFP, SODIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1ZSF
 
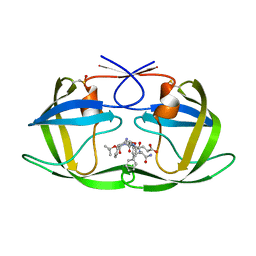 | | Crystal Structure of Complex of a Hydroxyethylamine Inhibitor with HIV-1 Protease at 2.0A Resolution | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-24 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the role of the R configuration of the reaction-intermediate isostere in HIV-1 protease-inhibitor binding: X-ray structure at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1ZPK
 
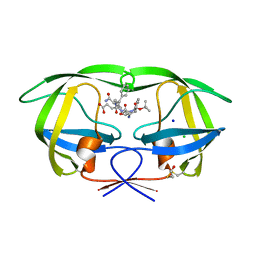 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[R-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | CHLORIDE ION, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN, ... | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational Study of Pseudopeptide Inhibitor Binding to HIV-1 Protease; Analysis of Four X-ray Structures
To be Published
|
|
1Z8C
 
 | | Crystal structure of the complex of mutant HIV-1 protease (l63P, A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[R-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN, SULFATE ION | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational Study of Pseudopeptide Inhibitor Binding to HIV-1 Protease; Analysis of Four X-ray Structures
To be Published
|
|
1ZBG
 
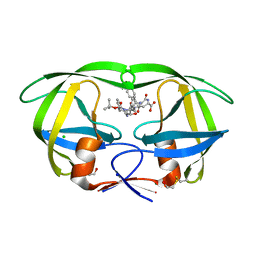 | | Crystal structure of a complex of mutant hiv-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[R-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | CHLORIDE ION, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN, ... | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-04-08 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Mutational Study of Pseudopeptide Inhibitor Binding to HIV-1 Protease; Analysis of Four X-ray Structures
To be Published
|
|
3RJA
 
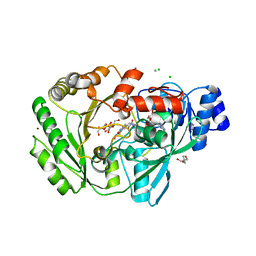 | | Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duskova, J, Skalova, T, Kolenko, P, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
3RJ8
 
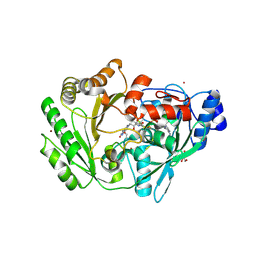 | | Crystal structure of carbohydrate oxidase from Microdochium nivale | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carbohydrate oxidase, ... | | Authors: | Duskova, J, Skalova, T, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Kolenko, P, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
6T3O
 
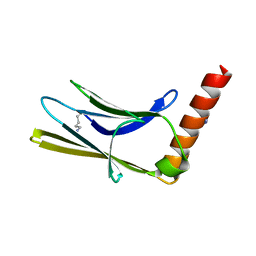 | | Crystal structure of the human myomesin domain 10 | | Descriptor: | AZIDE ION, Myomesin-1 | | Authors: | Duskova, J, Petrokova, H, Maly, P. | | Deposit date: | 2019-10-11 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Myomedin scaffold variants targeted to 10E8 HIV-1 broadly neutralizing antibody mimic gp41 epitope and elicit HIV-1 virus-neutralizing sera in mice.
Virulence, 12, 2021
|
|
1ZSR
 
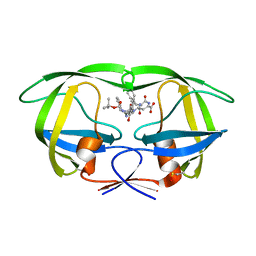 | | Crystal structure of wild type HIV-1 protease (BRU isolate) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-24 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the R configuration of the reaction-intermediate isostere in HIV-1 protease-inhibitor binding: X-ray structure at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2RGS
 
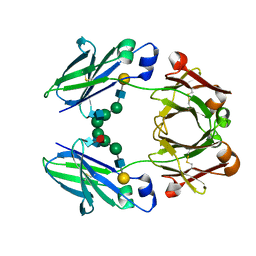 | | FC-fragment of monoclonal antibody IGG2B from Mus musculus | | Descriptor: | Ig gamma-2B heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J, Hasek, J. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | New insights into intra- and intermolecular interactions of immunoglobulins: crystal structure of mouse IgG2b-Fc at 2.1-A resolution
Immunology, 126, 2008
|
|
5J2S
 
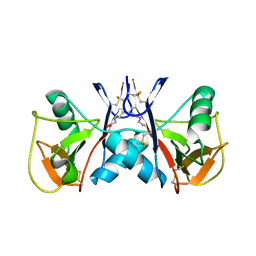 | | NKR-P1B from Rattus norvegicus | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1B allele A, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Skalova, T, Vanek, O, Blaha, J, Duskova, J, Hasek, J, Koval, T, Dohnalek, J. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of NKR-P1B from Rattus norvegicus
To Be Published
|
|
1IIQ
 
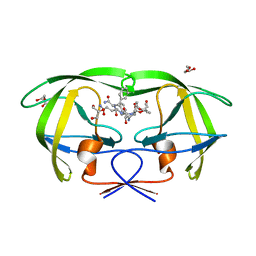 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE COMPLEXED WITH A HYDROXYETHYLAMINE PEPTIDOMIMETIC INHIBITOR | | Descriptor: | GLYCEROL, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Dohnalek, J, Hasek, J, Duskova, J, Petrokova, H, Hradilek, M, Soucek, M, Konvalinka, J, Brynda, J, Sedlacek, J, Fabry, M. | | Deposit date: | 2001-04-24 | | Release date: | 2002-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Hydroxyethylamine isostere of an HIV-1 protease inhibitor prefers its amine to the hydroxy group in binding to catalytic aspartates. A synchrotron study of HIV-1 protease in complex with a peptidomimetic inhibitor.
J.Med.Chem., 45, 2002
|
|
3CG8
 
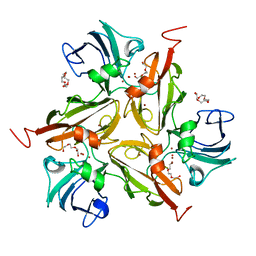 | | Laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, TETRAETHYLENE GLYCOL, ... | | Authors: | Skalova, T, Dohnalek, J, Ostergaard, L.H, Ostergaard, P.R, Kolenko, P, Duskova, J, Hasek, J. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | The Structure of the Small Laccase from Streptomyces coelicolor Reveals a Link between Laccases and Nitrite Reductases.
J.Mol.Biol., 385, 2009
|
|
1FQX
 
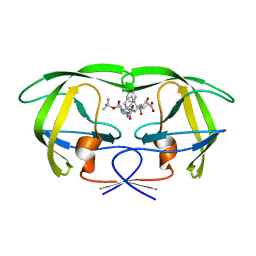 | | CRYSTAL STRUCTURE OF THE COMPLEX OF HIV-1 PROTEASE WITH A PEPTIDOMIMETIC INHIBITOR | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Dohnalek, J, Hasek, J, Duskova, J, Petrokova, H, Hradilek, M, Soucek, M, Konvalinka, J, Brynda, J, Sedlacek, J, Fabry, M. | | Deposit date: | 2000-09-07 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A distinct binding mode of a hydroxyethylamine isostere inhibitor of HIV-1 protease.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3KW8
 
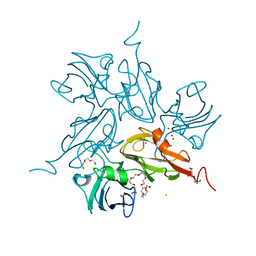 | | Two-domain laccase from Streptomyces coelicolor at 2.3 A resolution | | Descriptor: | COPPER (II) ION, FE (III) ION, Putative copper oxidase, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Duskova, J, Stepankova, A, Hasek, J, Ostergaard, L.H, Ostergaard, P.R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of laccase from Streptomyces coelicolor after soaking with potassium hexacyanoferrate and at an improved resolution of 2.3 A
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6I3L
 
 | | Bilirubin oxidase from Myrothecium verrucaria, mutant W396F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Koval, T, Svecova, L, Skalova, T, Kolenko, P, Duskova, J, Ostergaard, L.H, Dohnalek, J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trp-His covalent adduct in bilirubin oxidase is crucial for effective bilirubin binding but has a minor role in electron transfer.
Sci Rep, 9, 2019
|
|
8AQ8
 
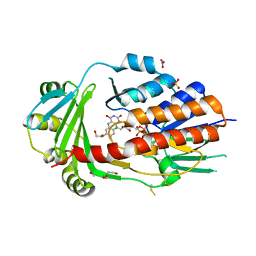 | | FAD-dependent monooxygenase from Stenotrophomonas maltophilia | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maly, M, Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetracycline-modifying enzyme SmTetX from Stenotrophomonas maltophilia.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6I3J
 
 | | Bilirubin oxidase from Myrothecium verrucaria in complex with ferricyanide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, CHLORIDE ION, ... | | Authors: | Svecova, L, Koval, T, Skalova, T, Kolenko, P, Duskova, J, Ostergaard, L.H, Dohnalek, J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Trp-His covalent adduct in bilirubin oxidase is crucial for effective bilirubin binding but has a minor role in electron transfer.
Sci Rep, 9, 2019
|
|
