7CZR
 
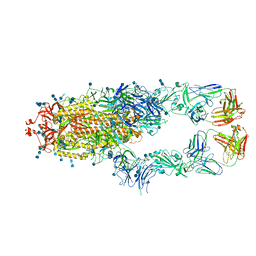 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZT
 
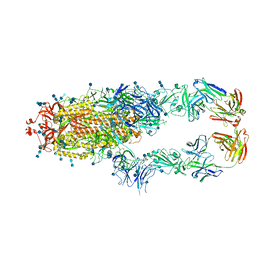 | | S protein of SARS-CoV-2 in complex bound with P5A-2G9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c689_light_IGLV5-37_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0D
 
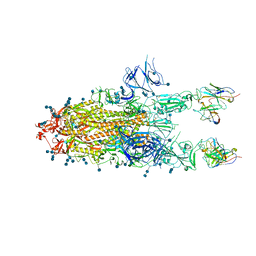 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | Authors: | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZS
 
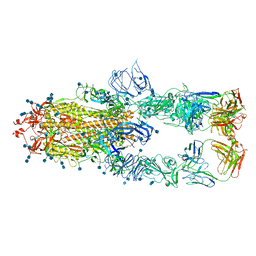 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZU
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-1B6_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy variable 3-30-3,Immunoglobulin heavy variable 3-30-3,Chain H of P5A-1B6_2B,Immunoglobulin gamma-1 heavy chain,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZV
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-1B6_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy variable 3-30-3,chain H of P5A-1B6_3B,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZX
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-1B9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c168_light_IGKV4-1_IGKJ4,Uncharacterized protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZY
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2F11_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZZ
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2F11_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0C
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3A1, ... | | Authors: | Yan, R.H, Wang, R.K, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0B
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_1B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | Authors: | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D00
 
 | | S protein of SARS-CoV-2 in complex bound with FabP5A-1B8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,chain H of FabP5A-1B8,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D03
 
 | | S protein of SARS-CoV-2 in complex bound with FabP5A-2G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZW
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7DMX
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
7DNA
 
 | |
2JE4
 
 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
2JG4
 
 | | Substrate-free IDE structure in its closed conformation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
6IJ7
 
 | | Crystal Structure of Arabidopsis thaliana UGT89C1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Rhamnosyltransferase protein | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
6IJD
 
 | | Crystal Structure of Arabidopsis thaliana UGT89C1 complexed with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP-glycosyltransferase 89C1 | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
6IJA
 
 | | Crystal Structure of Arabidopsis thaliana UGT89C1 complexed with UDP-L-rhamnose | | Descriptor: | UDP-glycosyltransferase 89C1, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
6J2R
 
 | | Crystal structure of Striga hermonthica HTL8 (ShHTL8) | | Descriptor: | GLYCEROL, Hyposensitive to light 8 | | Authors: | Zhang, Y.Y, Xi, Z. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
7JTP
 
 | | Crystal structure of Protac MS67 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | Elongin-B, Elongin-C, GLYCEROL, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
7JTO
 
 | | Crystal structure of Protac MS33 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(11-{[2-(4-{[4'-(4-methylpiperazin-1-yl)-3'-{[6-oxo-4-(trifluoromethyl)-5,6-dihydropyridine-3-carbonyl]amino}[1,1'-biphenyl]-3-yl]methyl}piperazin-1-yl)ethyl]amino}-11-oxoundecanoyl)-L-valyl-(4R)-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, Elongin-B, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
