5M6Y
 
 | | Cocrystal structure of cAMP-dependent Protein Kinase (PKA) in complex with a methylisoquinoline Fasudil-derivative | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)-4-methyl-isoquinoline, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2016-10-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Cocrystal structure of cAMP-dependent Protein Kinase (PKA) in complex with differently methylated Fasudil-derived ligands
To be Published
|
|
5M75
 
 | |
1E2S
 
 | | Crystal structure of an Arylsulfatase A mutant C69A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION, ... | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E3C
 
 | | Crystal structure of an Arylsulfatase A mutant C69S soaked in synthetic substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-13 | | Release date: | 2001-03-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E1Z
 
 | | Crystal structure of an Arylsulfatase A mutant C69S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E33
 
 | | Crystal structure of an Arylsulfatase A mutant P426L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-06 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defective oligomerization of arylsulfatase a as a cause of its instability in lysosomes and metachromatic leukodystrophy.
J. Biol. Chem., 277, 2002
|
|
1E6F
 
 | | Human MIR-receptor, repeat 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Rajashankar, K.R, Dauter, M, Dauter, Z, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4R02
 
 | | yCP in complex with BSc4999 (alpha-Keto Phenylamide) | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(2S,3S)-1-[(2,4-dimethylphenyl)amino]-2-hydroxy-5-methyl-1-oxohexan-3-yl}-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Voss, C, Scholz, C, Knorr, S, Beck, P, Stein, M, Zall, A, Kuckelkorn, U, Kloetzel, P.-M, Groll, M, Hamacher, K, Schmidt, B. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | alpha-Keto Phenylamides as P1'-Extended Proteasome Inhibitors.
Chemmedchem, 9, 2014
|
|
6YNC
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with the methylated Fasudil-derived fragment N-methylisoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-methylisoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YNB
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with short-chain Fasudil-derivative N-(2-aminoethyl)isoquinoline-5-sulfonamide (soaked) | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)ISOQUINOLINE-5-SULFONAMIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6Y0B
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with quinazolin-4-amine and PKI (5-24) | | Descriptor: | DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
6Y05
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with adenine and PKI (5-24) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENINE, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
7AXV
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with 5-isoquinolinesulfonic acid and PKI (5-24) | | Descriptor: | DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
7AXT
 
 | |
7AXW
 
 | |
1FD9
 
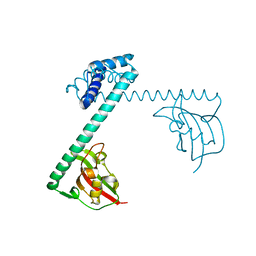 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | Authors: | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
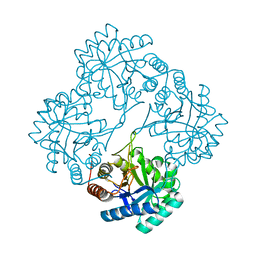
6U57
 
 | |
7BET
 
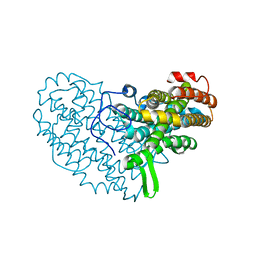 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
6Y2O
 
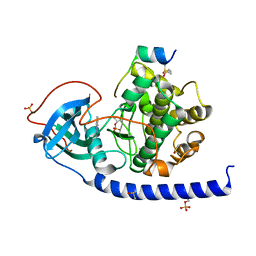 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with 1,7-Naphthyridin-8-amine and PKI (5-24) | | Descriptor: | 1,7-naphthyridin-8-amine, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
6YNT
 
 | | Crystal structure of the cAMP-dependent protein kinase A in complex with aminofasudil and PKI (5-24) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)isoquinolin-1-amine, ... | | Authors: | Oebbeke, M, Gerber, H.-D, Heine, A, Klebe, G. | | Deposit date: | 2020-04-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6Y8C
 
 | |
7AT6
 
 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
3QO6
 
 | | Crystal structure analysis of the plant protease Deg1 | | Descriptor: | Protease Do-like 1, chloroplastic, peptide | | Authors: | Clausen, T. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6SQ1
 
 | | Crystal structure of cAMP-dependent Protein Kinase A (CHO PKA) in complex with Aminofasudil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)isoquinolin-1-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A crystallographic study of cAMP-dependent Protein Kinase A in complex with different Fasudil-derivatives
To Be Published
|
|
1Y1E
 
 | | human formylglycine generating enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-alpha-formyglycine-generating enzyme, CALCIUM ION | | Authors: | Rudolph, M.G, Dickmanns, A, Ficner, R. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme.
Cell(Cambridge,Mass.), 121, 2005
|
|
