3M2W
 
 | | Crystal structure of MAPKAK kinase 2 (MK2) complexed with a spiroazetidine-tetracyclic ATP site inhibitor | | Descriptor: | 2'-(2-fluorophenyl)-1-methyl-6',8',9',11'-tetrahydrospiro[azetidine-3,10'-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin]-7'(5'H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Scheufler, C. | | Deposit date: | 2010-03-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | In vivo and in vitro SAR of tetracyclic MAPKAP-K2 (MK2) inhibitors. Part II.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KGA
 
 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a potent 3-aminopyrazole ATP site inhibitor | | Descriptor: | 6-{3-amino-1-[3-(1H-indol-6-yl)phenyl]-1H-pyrazol-4-yl}-3,4-dihydroisoquinolin-1(2H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Velcicky, J, Izaac, A, Be, C, Huppertz, C, Pflieger, D, Schlapbach, A, Scheufler, C. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel 3-aminopyrazole inhibitors of MK-2 discovered by scaffold hopping strategy.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M42
 
 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a tetracyclic ATP site inhibitor | | Descriptor: | 2-[5-(2-methoxyethoxy)pyridin-3-yl]-8,9,10,11-tetrahydro-7H-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin-7-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Scheufler, C, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Kroemer, M. | | Deposit date: | 2010-03-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel pyrrolo[2,3-f]isoquinoline based MAPKAP-K2 (MK2) inhibitors with potent in vitro and in vivo activity
To be Published
|
|
6Z10
 
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-chloranyl-2-fluoranyl-5-[(3~{R})-piperidin-3-yl]oxy-phenyl]-2-fluoranyl-phenyl]carbonylamino]-5-fluoranyl-phenyl]ethanoic acid, CHLORIDE ION, ... | | Authors: | Haffke, M, Villard, F. | | Deposit date: | 2020-05-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Discovery and Optimization of Novel SUCNR1 Inhibitors: Design of Zwitterionic Derivatives with a Salt Bridge for the Improvement of Oral Exposure.
J.Med.Chem., 63, 2020
|
|
6GGN
 
 | | In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor | | Descriptor: | (4~{S})-4-(4-chloranyl-2-methyl-phenyl)-5-(5-chloranyl-2-methyl-phenyl)-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Kallen, J. | | Deposit date: | 2018-05-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5LN2
 
 | |
7AK0
 
 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-[4-[4-(aminomethyl)pyrazol-1-yl]-3-chloranyl-phenyl]-3-[(3~{R})-6-bromanyl-3,4-dihydro-2~{H}-chromen-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
7AK1
 
 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-(3-chloranyl-4-methoxy-phenyl)-3-[7-[(3~{S})-3-(methoxymethyl)morpholin-4-yl]-2-methyl-pyrazolo[1,5-a]pyrimidin-6-yl]urea, MAGNESIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
6H4A
 
 | | Human MALT1(329-728) in complex with MLT-748 | | Descriptor: | 1-[2-chloranyl-7-[(1~{R},2~{R})-1,2-dimethoxypropyl]pyrazolo[1,5-a]pyrimidin-6-yl]-3-[5-chloranyl-6-(1,2,3-triazol-2-yl)pyridin-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M, Renatus, M. | | Deposit date: | 2018-07-20 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient.
Nat. Chem. Biol., 15, 2019
|
|
6YN9
 
 | | MALT1(329-728) in complex with a sulfonamide containing compound | | Descriptor: | 5-[4-[(2,6-diethylphenyl)sulfamoyl]-3-methyl-phenyl]furan-3-carboxylic acid, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
6F7I
 
 | | human MALT1(329-728) IN COMPLEX WITH MLT-747 | | Descriptor: | 1-[2-chloranyl-7-[(1~{S})-1-methoxyethyl]pyrazolo[1,5-a]pyrimidin-6-yl]-3-(5-chloranyl-6-pyrrolidin-1-ylcarbonyl-pyridin-3-yl)urea, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Renatus, M, Renatus, M. | | Deposit date: | 2017-12-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient.
Nat. Chem. Biol., 15, 2019
|
|
6YN8
 
 | | Human MALT1(334-719) in complex with a tetrazole containing compound | | Descriptor: | 3-azanyl-3-methyl-~{N}-[(3~{R})-4-oxidanylidene-5-[[4-[2-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]-2,3-dihydro-1,5-benzoxazepin-3-yl]butanamide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
9H4D
 
 | | Crystal structure of IL-17A in complex with compound 18 | | Descriptor: | (E)-N-[(1S)-1-[4,4-bis(fluoranyl)cyclohexyl]-2-oxidanylidene-2-[[5-[(2-oxidanylidene-3H-benzimidazol-1-yl)methyl]-1,3-thiazol-2-yl]amino]ethyl]-3-cyclopropyl-2-fluoranyl-prop-2-enamide, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-10-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Thiazole-Based IL-17 Inhibitors Discovered by Scaffold Morphing.
Chemmedchem, 20, 2025
|
|
9H4O
 
 | | Crystal structure of IL-17A in complex with compound 11 | | Descriptor: | (E)-N-[(1S)-1-[4,4-bis(fluoranyl)cyclohexyl]-2-oxidanylidene-2-[[5-[[(4S)-2-oxidanylidene-4-(trifluoromethyl)imidazolidin-1-yl]methyl]-1,3-thiazol-2-yl]amino]ethyl]-3-cyclopropyl-2-fluoranyl-prop-2-enamide, Interleukin-17A, SULFATE ION | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-10-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thiazole-Based IL-17 Inhibitors Discovered by Scaffold Morphing.
Chemmedchem, 20, 2025
|
|
9FKX
 
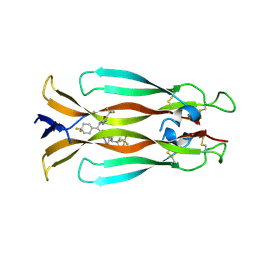 | | Crystal structure of IL-17A in complex with compound 18 | | Descriptor: | (4~{S})-1-[(~{R})-[2-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[[5-[1,1-bis(fluoranyl)ethyl]-1,3,4-oxadiazol-2-yl]amino]methyl]imidazo[1,2-b]pyridazin-7-yl]-cyclopropyl-methyl]-4-(trifluoromethyl)imidazolidin-2-one, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9FL3
 
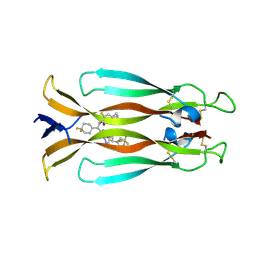 | | Crystal structure of IL-17A in complex with compound 26 | | Descriptor: | (~{E})-~{N}-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[7-[(1~{S})-2-methoxy-1-[(4~{S})-2-oxidanylidene-4-(trifluoromethyl)imidazolidin-1-yl]ethyl]imidazo[1,2-b]pyridazin-2-yl]methyl]-3-cyclopropyl-2-fluoranyl-prop-2-enamide, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 67, 2024
|
|
6Q9L
 
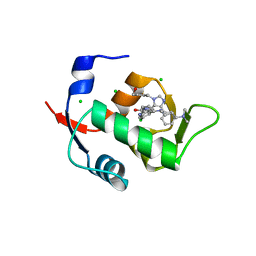 | |
6Q9U
 
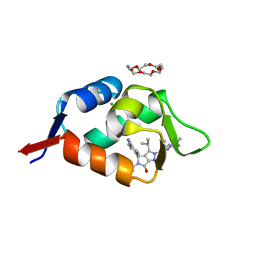 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 12 AT 2.4A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-[(4~{S})-5-(5-chloranyl-2-oxidanylidene-1~{H}-pyridin-3-yl)-2-[2-(dimethylamino)-4-methoxy-pyrimidin-5-yl]-6-oxidanylidene-3-propan-2-yl-4~{H}-pyrrolo[3,4-c]pyrazol-4-yl]benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9O
 
 | |
6Q9Q
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 13 AT 2.1A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-(4-cyclohexylpiperazin-1-yl)ethyl]-1~{H}-indole-2-carboxamide, GLYCEROL, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9W
 
 | | X-ray structure of compound 15 bound to HdmX: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | (4~{S})-4-(4-chlorophenyl)-5-[(1~{S})-1-(3-chlorophenyl)ethyl]-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9H
 
 | |
6Q9Y
 
 | |
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q96
 
 | |
