4MUT
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUS
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUR
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3D1R
 
 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
4MUQ
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.364 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N3O
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni. | | Descriptor: | CALCIUM ION, Putative D-glycero-D-manno-heptose 7-phosphate kinase | | Authors: | Minasov, G, Wawrzak, Z, Gordon, E, Onopriyenko, O, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-07 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4NVR
 
 | | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Stam, J, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica.
TO BE PUBLISHED
|
|
4OMV
 
 | | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4 | | Descriptor: | Macrophage growth locus, subunit A | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4
TO BE PUBLISHED
|
|
4OHN
 
 | | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, HISTIDINE | | Authors: | Brunzelle, J.S, Wawrzak, W, Yim, Y, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
To be Published
|
|
4OAK
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine-D-Alanine and copper (II) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OFX
 
 | | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii | | Descriptor: | Cystathionine beta-synthase, SODIUM ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii
To be Published
|
|
4O96
 
 | | 2.60 Angstrom resolution crystal structure of a protein kinase domain of type III effector NleH2 (ECs1814) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, type III effector protein kinase | | Authors: | Anderson, S.M, Halavaty, A.S, Wawrzak, Z, Kudritska, M, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type III Effector NleH2 from Escherichia coli O157:H7 str. Sakai Features an Atypical Protein Kinase Domain.
Biochemistry, 53, 2014
|
|
4OEN
 
 | | Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | ACETATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A
To be Published
|
|
4OC9
 
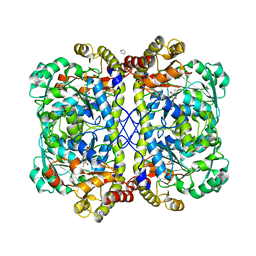 | | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205 | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205
To be Published
|
|
2RC3
 
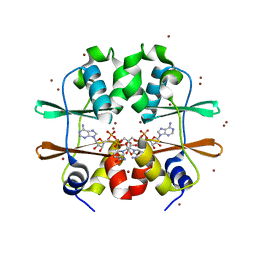 | | Crystal structure of CBS domain, NE2398 | | Descriptor: | BROMIDE ION, CBS domain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dong, A, Xu, X, Korniyenko, Y, Yakunin, A, Zheng, H, Walker, J.R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CBS domain, NE2398.
To be Published
|
|
3BRM
 
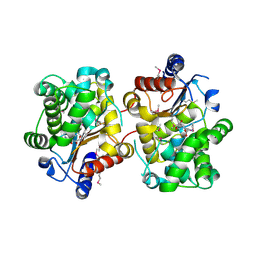 | | Crystal structure of the covalent complex between the Bacillus subtilis glutaminase YbgJ and 5-oxo-L-norleucine formed by reaction of the protein with 6-diazo-5-oxo-L-norleucine | | Descriptor: | 5-OXO-L-NORLEUCINE, Glutaminase 1 | | Authors: | Singer, A.U, Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2HS5
 
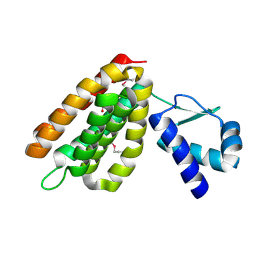 | | Structural Genomics, the crystal structure of a putative transcriptional regulator GntR from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, putative transcriptional regulator GntR | | Authors: | Tan, K, Skarina, T, Onopriyenko, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-21 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a putative transcriptional regulator GntR from Rhodococcus sp. RHA1
To be Published
|
|
3D7N
 
 | | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens | | Descriptor: | Flavodoxin, WrbA-like protein | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens
To be Published, 2008
|
|
3DCI
 
 | | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Arylesterase, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Binkowski, T.A, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58
TO BE PUBLISHED
|
|
3DED
 
 | | C-terminal domain of Probable hemolysin from Chromobacterium violaceum | | Descriptor: | CALCIUM ION, Probable hemolysin | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of C-terminal domain of Probable hemolysin from Chromobacterium violaceum
To be Published
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
3DNH
 
 | | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58 | | Descriptor: | uncharacterized protein Atu2129 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58
To be Published
|
|
3DRW
 
 | | Crystal Structure of a Phosphofructokinase from Pyrococcus horikoshii OT3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-specific phosphofructokinase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Kochinyan, S, Brown, G, Cuff, M.E, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Jia, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
3EFE
 
 | | The crystal structure of the thiJ/pfpI family protein from Bacillus anthracis | | Descriptor: | SULFATE ION, ThiJ/pfpI family protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the thiJ/pfpI family protein from Bacillus anthracis
To be Published
|
|
3E9Q
 
 | | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-23 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri
To be Published
|
|
