4Y2G
 
 | |
4YNH
 
 | |
4YV4
 
 | |
5AJI
 
 | | MscS D67R1 high resolution | | Descriptor: | DECANE, HEXANE, N-OCTANE, ... | | Authors: | Naismith, J.H, Pliotas, C. | | Deposit date: | 2015-02-24 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Role of Lipids in Mechanosensation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5A2F
 
 | | Two membrane distal IgSF domains of CD166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD166 ANTIGEN | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5A2E
 
 | | Extracellular SRCR domains of human CD6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-CELL DIFFERENTIATION ANTIGEN CD6 | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IS4
 
 | |
4JML
 
 | |
2W08
 
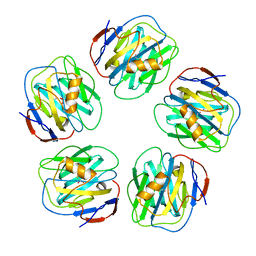 | | The structure of serum amyloid P component bound to 0-phospho- threonine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHOTHREONINE, ... | | Authors: | Kolstoe, S.E, Pepys, M.B, Wood, S.P. | | Deposit date: | 2008-08-12 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Dissection of Alzheimer'S Disease Neuropathology by Depletion of Serum Amyloid P Component.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WQJ
 
 | |
2WQI
 
 | |
2WTT
 
 | |
2XWR
 
 | |
2Y3V
 
 | | N-terminal head domain of Danio rerio SAS-6 | | Descriptor: | SPINDLE ASSEMBLY ABNORMAL PROTEIN 6 HOMOLOG | | Authors: | van Breugel, M. | | Deposit date: | 2010-12-27 | | Release date: | 2011-02-09 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of SAS-6 suggest its organization in centrioles.
Science, 331, 2011
|
|
2Y3W
 
 | |
2YBG
 
 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ZQQ
 
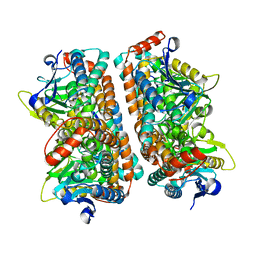 | | Crystal structure of human AUH (3-methylglutaconyl-coa hydratase) mixed with (AUUU)24A RNA | | Descriptor: | Methylglutaconyl-CoA hydratase | | Authors: | Kurimoto, K, Kuwasako, K, Muto, Y, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AU-rich RNA-binding induces changes in the quaternary structure of AUH
Proteins, 75, 2009
|
|
2ZQR
 
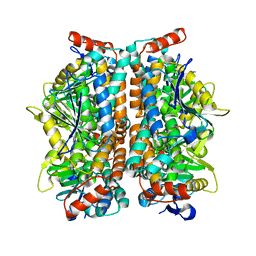 | | Crystal structure of AUH without RNA | | Descriptor: | Methylglutaconyl-CoA hydratase | | Authors: | Kurimoto, K, Kuwasako, K, Muto, Y, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AU-rich RNA-binding induces changes in the quaternary structure of AUH
Proteins, 75, 2009
|
|
1JNJ
 
 | | NMR solution structure of the human beta2-microglobulin | | Descriptor: | beta2-microglobulin | | Authors: | Verdone, G, Corazza, A, Viglino, P, Pettirossi, F, Giorgetti, S, Mangione, P, Andreola, A, Stoppini, M, Bellotti, V, Esposito, G. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human beta2-microglobulin reveals the prodromes of its amyloid transition.
Protein Sci., 11, 2002
|
|
1LOZ
 
 | |
1LYY
 
 | |
6ELU
 
 | |
6EOJ
 
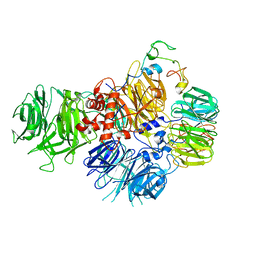 | | PolyA polymerase module of the cleavage and polyadenylation factor (CPF) from Saccharomyces cerevisiae | | Descriptor: | Polyadenylation factor subunit 2,Polyadenylation factor subunit 2, Protein CFT1, ZINC ION, ... | | Authors: | Casanal, A, Kumar, A, Hill, C.H, Emsley, P, Passmore, L. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-15 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Architecture of eukaryotic mRNA 3'-end processing machinery.
Science, 358, 2017
|
|
