4M2O
 
 | |
4M2P
 
 | |
4MLW
 
 | |
4YI8
 
 | |
4YI9
 
 | |
4M2Q
 
 | | Crystal structure of non-myristoylated recoverin with Cysteine-39 oxidized to sulfenic acid | | Descriptor: | CALCIUM ION, Recoverin | | Authors: | Prem Kumar, R, Chakrabarti, K, Kern, D, Oprian, D.D. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Highly Conserved Cysteine of Neuronal Calcium-sensing Proteins Controls Cooperative Binding of Ca2+ to Recoverin.
J.Biol.Chem., 288, 2013
|
|
7M05
 
 | |
6V0N
 
 | | PRMT5 bound to PBM peptide from Riok1 | | Descriptor: | Methylosome protein 50, Protein arginine N-methyltransferase 5, Riok1 PBM peptide, ... | | Authors: | McMIllan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
6V0P
 
 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
6V0O
 
 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
7L27
 
 | |
7L28
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | Descriptor: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-12-16 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
 | |
7KWE
 
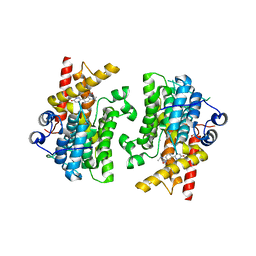 | | Crystal structure of the catalytic domain of human PDE3A bound to DNMDP | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRD
 
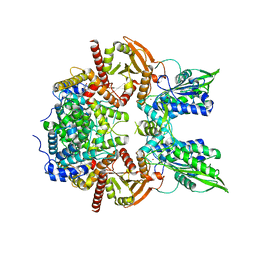 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRE
 
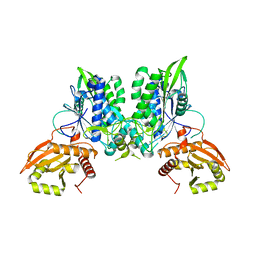 | |
7LRC
 
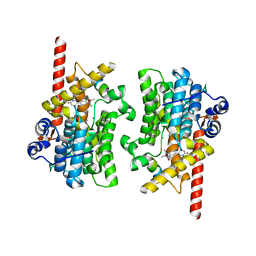 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
