3ICV
 
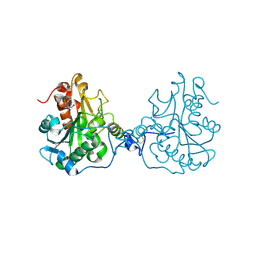 | | Structural Consequences of a Circular Permutation on Lipase B from Candida Antartica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3ICW
 
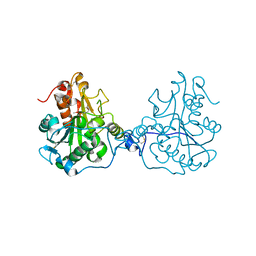 | | Structure of a Circular Permutation on Lipase B from Candida Antartica with Bound Suicide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, PHOSPHATE ION, ... | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S.A, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
5VST
 
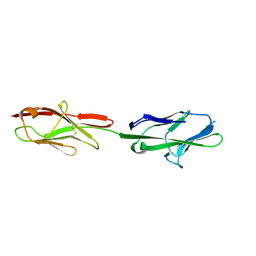 | | Crystal structure of murine CEACAM1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Biliary glycoprotein | | Authors: | Peng, G, Yang, Y, Pasquarella, J.R, Xu, L, Qian, Z, Holmes, K.V, Li, F. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism for coronavirus-driven evolution of mouse receptor
J. Biol. Chem., 292, 2017
|
|
3UGC
 
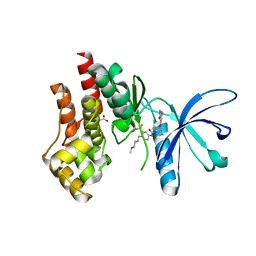 | | Structural basis of Jak2 inhibition by the type II inhibtor NVP-BBT594 | | Descriptor: | 5-{[6-(acetylamino)pyrimidin-4-yl]oxy}-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-2,3-dihydro-1H-indole-1-carboxamide, MALONATE ION, Tyrosine-protein kinase JAK2 | | Authors: | Scheufler, C, Tavares, G.A, Manley, P.W, Pissot-Soldermann, C, Kroemer, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent.
Cancer Discov, 2, 2012
|
|
3R4D
 
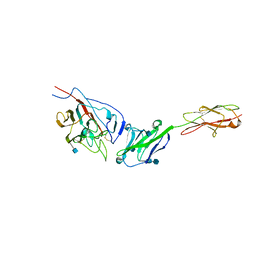 | | Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEA-related cell adhesion molecule 1, ... | | Authors: | Peng, G.Q, Sun, D.W, Rajashankar, K.R, Qian, Z.H, Holmes, K.V, Li, F. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5KWB
 
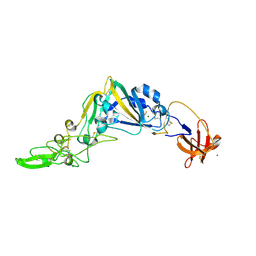 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
5GNB
 
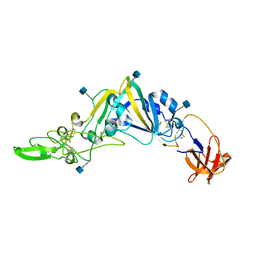 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
6AKW
 
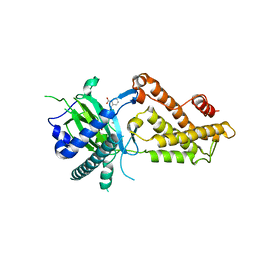 | | Crystal structure of RNA dioxygenase bound with an inhibitor | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2018-09-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia.
Cancer Cell, 35, 2019
|
|
7X6L
 
 | |
7X6O
 
 | |
7X9Q
 
 | | Crystal structure of human STING complexed with compound BSP16 | | Descriptor: | (2R)-4-(5,6-dimethoxy-1-benzoselenophen-2-yl)-2-ethyl-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Selenium-Containing STING Agonists as Orally Available Antitumor Agents.
J.Med.Chem., 65, 2022
|
|
6M6X
 
 | | Oridonin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6M60
 
 | | Plumbagin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (2~{R})-2-methyl-5-oxidanyl-2,3-dihydronaphthalene-1,4-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
7V9U
 
 | | Cryo-EM structure of E.coli retron-Ec86 (RT-msDNA-RNA) at 3.2 angstrom | | Descriptor: | DNA (105-MER), RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*G)-3'), RNA (81-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
7XJG
 
 | | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | | Descriptor: | DNA (105-MER), MAGNESIUM ION, RNA (14-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2022-04-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
7DBG
 
 | | Yeast CRM1e (apo) in complex with Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-10-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
8GWH
 
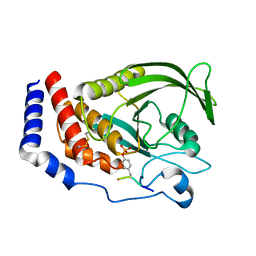 | |
8GVL
 
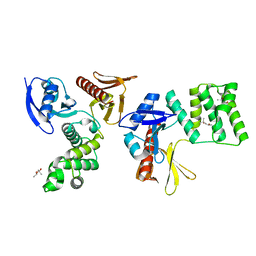 | | PTPN21 FERM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GVV
 
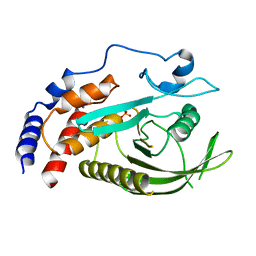 | | PTPN21 PTP domain C1108S mutant | | Descriptor: | IODIDE ION, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GXE
 
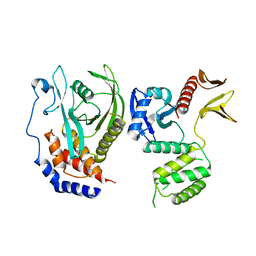 | | PTPN21 FERM PTP complex | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
7X9P
 
 | | Crystal structure of human STING complexed with compound BSP17 | | Descriptor: | 4-[6-methoxy-5-[3-[[6-methoxy-2-(4-oxidanyl-4-oxidanylidene-butanoyl)-1-benzoselenophen-5-yl]oxy]propoxy]-1-benzoselenophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selenium-containing STING agonists as orally available anti tumor agents
To be published
|
|
