4XZI
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0049 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,4-dioxo-3-(2,3,4,5-tetrabromo-6-methoxybenzyl)-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZN
 
 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZL
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
1RNA
 
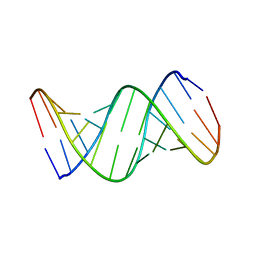 | | CRYSTALLOGRAPHIC STRUCTURE OF AN RNA HELIX: [U(U-A)6A]2 | | Descriptor: | RNA (5'-R(*UP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*A)-3') | | Authors: | Dock-Bregeon, A.C, Chevrier, B, Podjarny, A, Johnson, J, De Bear, J.S, Gough, G.R, Gilham, P.T, Moras, D. | | Deposit date: | 1990-02-01 | | Release date: | 1991-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic structure of an RNA helix: [U(UA)6A]2.
J.Mol.Biol., 209, 1989
|
|
3GHU
 
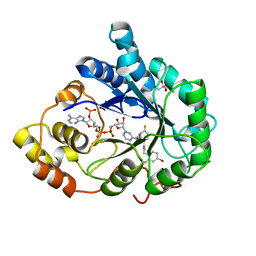 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Forth stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHS
 
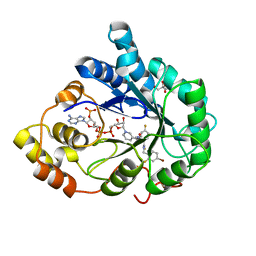 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Second stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHR
 
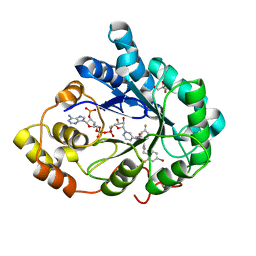 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. First stage of radiation damage | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHT
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Third stage of radiation damage. | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
5NRA
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7g | | Descriptor: | 1-(5-azanyl-4~{H}-1,2,4-triazol-3-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-(2-methylpropyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
5OUJ
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 39 | | Descriptor: | 2-[(1~{R})-5-(4-chlorophenyl)-9-fluoranyl-3-methyl-1-oxidanyl-1~{H}-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OUK
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 41 | | Descriptor: | 2-[9-fluoranyl-5-(4-methoxyphenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OU0
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
1US0
 
 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | Authors: | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | Deposit date: | 2003-11-16 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.66 Å) | | Cite: | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
2AGT
 
 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
3MU4
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU0
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU8
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU5
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MTY
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU1
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1X97
 
 | | Crystal structure of Aldose Reductase complexed with 2R4S (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2R,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X96
 
 | | Crystal structure of Aldose Reductase with citrates bound in the active site | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X98
 
 | | Crystal structure of Aldose Reductase complexed with 2S4R (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2S,4R)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, CITRIC ACID, ... | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1Z8A
 
 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
