2HN2
 
 | |
2HN1
 
 | |
2F6U
 
 | |
3RVY
 
 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.7 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3RVZ
 
 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.8 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3RW0
 
 | | Crystal structure of the NavAb voltage-gated sodium channel (Met221Cys, 2.95 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
4EKW
 
 | | Crystal structure of the NavAb voltage-gated sodium channel (wild-type, 3.2 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein, PHOSPHATE ION | | Authors: | Payandeh, J, Gamal El-Din, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2012-04-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of a voltage-gated sodium channel in two potentially inactivated states.
Nature, 486, 2012
|
|
5BO1
 
 | |
7SEL
 
 | | E. coli MsbA in complex with LPS and inhibitor G7090 (compound 3) | | Descriptor: | (2E)-3-{7-[(1S)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-1-methylnaphthalen-2-yl}prop-2-enoic acid, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Payandeh, J, Koth, C.M, Verma, V.A. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.978 Å) | | Cite: | Discovery of Inhibitors of the Lipopolysaccharide Transporter MsbA: From a Screening Hit to Potent Wild-Type Gram-Negative Activity.
J.Med.Chem., 65, 2022
|
|
6XLP
 
 | | Structure of the essential inner membrane lipopolysaccharide-PbgA complex | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-3-O-[(1R,3R)-1,3-dihydroxytetradecyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-1-O-phosphono-alpha-D-glucopyranose-(6-1)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)]1,5-anhydro-2-deoxy-2-{[(1S,3R)-1-hydroxy-3-(pentanoyloxy)undecyl]amino}-4-O-phosphono-D-glucitol, ... | | Authors: | Payandeh, J, Clairefeuille, T. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the essential inner membrane lipopolysaccharide-PbgA complex.
Nature, 584, 2020
|
|
7JWQ
 
 | |
7JWP
 
 | | Fab CJ11 in complex IL-18 peptide liberated by Caspase cleavage | | Descriptor: | Fab CJ11 Heavy chain, Fab CJ11 Light chain, IL-18 peptide | | Authors: | Payandeh, J, Ho, H. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of a caspase cleavage motif antibody reveals insights into noncanonical inflammasome function.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7U8G
 
 | | Cryo-EM structure of the core human NADPH oxidase NOX2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7G5 - heavy chain, ... | | Authors: | Noreng, S, Ota, N, Sun, Y, Masureel, M, Payandeh, J, Yi, T, Koerber, J.T. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the core human NADPH oxidase NOX2.
Nat Commun, 13, 2022
|
|
5EK0
 
 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
4OH3
 
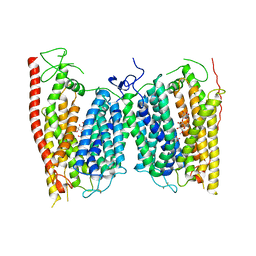 | | Crystal structure of a nitrate transporter | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, NITRATE ION, Nitrate transporter 1.1 | | Authors: | Sun, J, Bankston, J.R, Payandeh, J, Hinds, T.R, Zagotta, W.N, Zheng, N. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the plant dual-affinity nitrate transporter NRT1.1.
Nature, 507, 2014
|
|
8F0Q
 
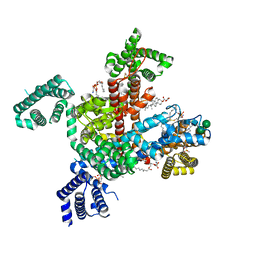 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4-yl}methoxy)-2-fluoro-N-(methanesulfonyl)benzamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0S
 
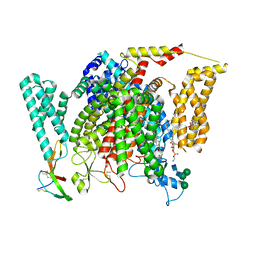 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0P
 
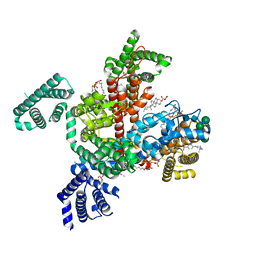 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0R
 
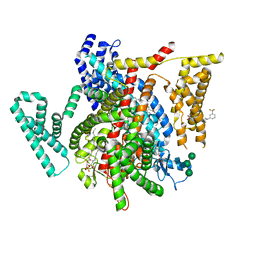 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
6BPL
 
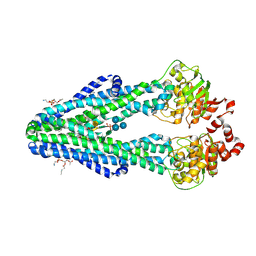 | | E. coli MsbA in complex with LPS and inhibitor G907 | | Descriptor: | (2E)-3-{6-[(1S)-1-(2-chloro-6-cyclopropylphenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-amino-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
6BPP
 
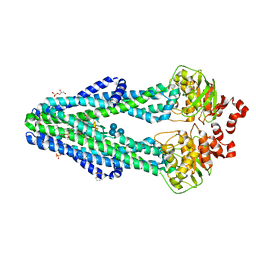 | | E. coli MsbA in complex with LPS and inhibitor G092 | | Descriptor: | (2E)-3-{6-[(1S)-1-(3-amino-2,6-dichlorophenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
9CNW
 
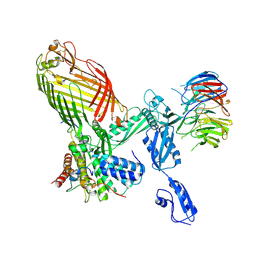 | | CryoEM structure of the APO-BAM complex in DDM detergent | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CNY
 
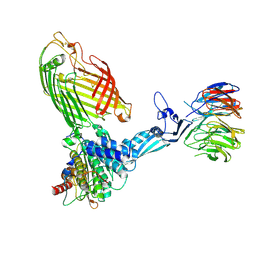 | | CryoEM structure of the APO-BAM complex in SMA nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CNZ
 
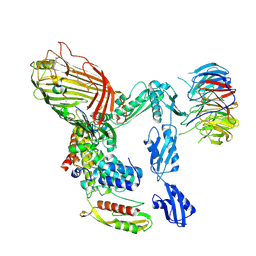 | | Structure of BAM complexed with PTB2 ligand in detergent | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CO0
 
 | | CryoEM structure of BAM in complex with the PTB2 open-state inhibitor (in SMA nanodisc) | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
