6XLP
 
 | | Structure of the essential inner membrane lipopolysaccharide-PbgA complex | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-3-O-[(1R,3R)-1,3-dihydroxytetradecyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-1-O-phosphono-alpha-D-glucopyranose-(6-1)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)]1,5-anhydro-2-deoxy-2-{[(1S,3R)-1-hydroxy-3-(pentanoyloxy)undecyl]amino}-4-O-phosphono-D-glucitol, ... | | 著者 | Payandeh, J, Clairefeuille, T. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the essential inner membrane lipopolysaccharide-PbgA complex.
Nature, 584, 2020
|
|
2CKX
 
 | |
6IG6
 
 | |
6IG7
 
 | |
4LW4
 
 | |
4LW2
 
 | |
6JLD
 
 | |
6JL9
 
 | |
8GTI
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | 分子名称: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | 著者 | Cho, H.S, Kim, H. | | 登録日 | 2022-09-08 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | 分子名称: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | 著者 | Cho, H.S, Kim, H. | | 登録日 | 2022-09-08 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTG
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | 分子名称: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | 著者 | Cho, H.S, Kim, H. | | 登録日 | 2022-09-08 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
7WKR
 
 | |
7WUC
 
 | |
6L18
 
 | |
4RDI
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | 著者 | Park, S.Y, Kim, S, Lee, H. | | 登録日 | 2014-09-19 | | 公開日 | 2015-08-05 | | 最終更新日 | 2015-10-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
4RDH
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | 分子名称: | ADENOSINE MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | 著者 | Park, S.Y, Kim, S, Lee, H. | | 登録日 | 2014-09-19 | | 公開日 | 2015-08-05 | | 最終更新日 | 2015-10-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
7YQ4
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ6
 
 | | human insulin receptor bound with A62 DNA aptamer | | 分子名称: | IR-A62 aptamer, Isoform Short of Insulin receptor | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ3
 
 | | human insulin receptor bound with A43 DNA aptamer and insulin | | 分子名称: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7SVL
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-2 | | 分子名称: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 9 | | 著者 | Lammens, A, Hollenstein, K, Klein, D.J. | | 登録日 | 2021-11-19 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVM
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-2 | | 分子名称: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 8, trimethylamine oxide | | 著者 | Lammens, A, Hollenstein, K, Klein, D.J. | | 登録日 | 2021-11-19 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | 分子名称: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | 著者 | Lammens, A, Hollenstein, K, Klein, D.J. | | 登録日 | 2021-11-19 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVO
 
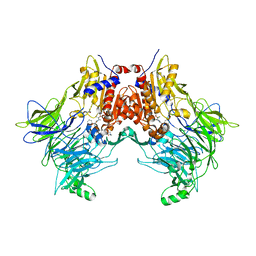 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | 分子名称: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | 著者 | Lammens, A, Hollenstein, K, Klein, D.J. | | 登録日 | 2021-11-19 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
5KUT
 
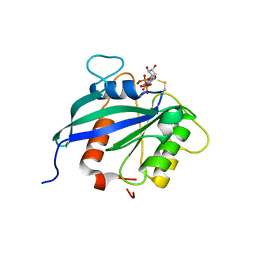 | | hMiro2 C-terminal GTPase domain, GDP-bound | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | 著者 | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | 登録日 | 2016-07-13 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.693 Å) | | 主引用文献 | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
