7ODF
 
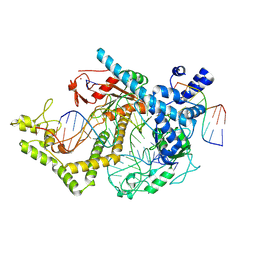 | | Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3 | | Descriptor: | Cas_phi3, DNA (5'-D(P*GP*G)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*TP*CP*AP*G)-3'), ... | | Authors: | Carabias del Rey, A, Fugilsang, A, Temperini, P, Pape, T, Sofos, N, Stella, S, Erledsson, S, Montoya, G. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.
Nat Commun, 12, 2021
|
|
7AF2
 
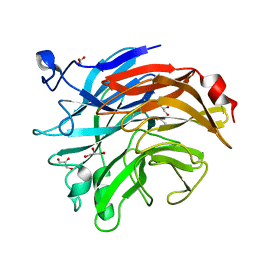 | | Salmonella typhimurium neuraminidase mutant (D62G) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.792 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
7AEY
 
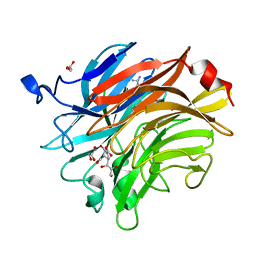 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | Descriptor: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
1W7Z
 
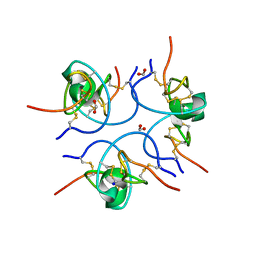 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | Descriptor: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | Deposit date: | 2004-09-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The molecular basis for cocaine binding to the human dopamine transporter
To Be Published
|
|
1ML5
 
 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | Deposit date: | 2002-08-30 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
1OIJ
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PUTATIVE ALKYLSULFATASE ATSK, SODIUM ION | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OIK
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with fe, alphaketoglutarate and 2-ethyl-1-hexanesulfuric acid | | Descriptor: | (2R)-2-ETHYL-1-HEXANESULFONIC ACID, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OII
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with iron and alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OIH
 
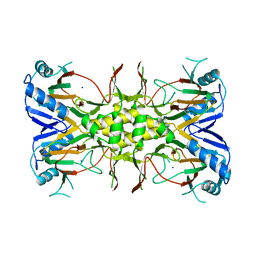 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase | | Descriptor: | PUTATIVE ALKYLSULFATASE ATSK, SODIUM ION | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) -Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
8AXB
 
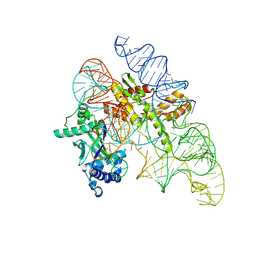 | | Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, sgRNA | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Molina, R, Pape, T, Lopez-Mendez, B, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural prediction of the Cas12k interaction with the transposon pre-integration complex
To Be Published
|
|
8AXA
 
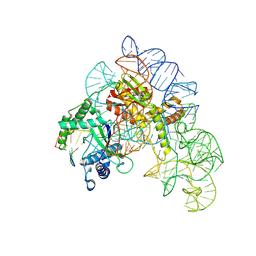 | | Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, DNA non-target strand, DNA target strand, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Fuglsang, A, Pape, T, Mesa, P, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
6S6B
 
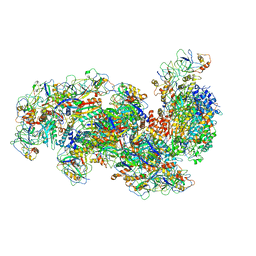 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8B
 
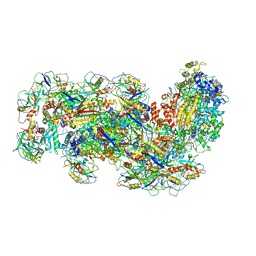 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
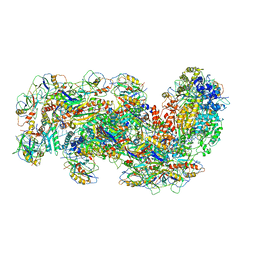 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8BJF
 
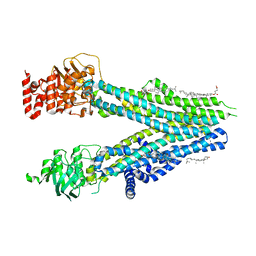 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (inward-facing conformation) | | Descriptor: | ATP-binding cassette sub-family C member 4, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWQ
 
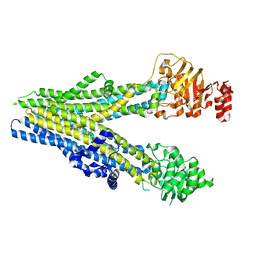 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with topotecan) | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWP
 
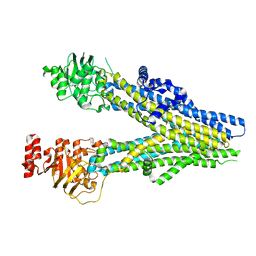 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with methotrexate) | | Descriptor: | ATP-binding cassette sub-family C member 4, METHOTREXATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWO
 
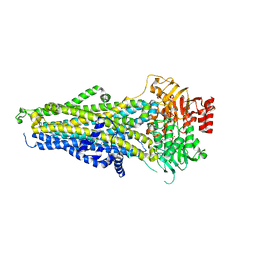 | | Cryo-EM structure of nanodisc-reconstituted human MRP4 with E1202Q mutation (outward-facing occluded conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWR
 
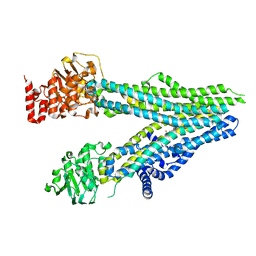 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with prostaglandin E2) | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
6SIC
 
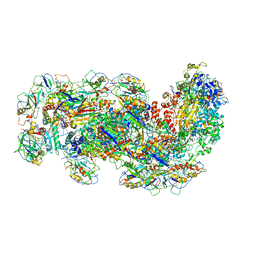 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8E
 
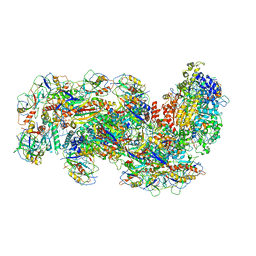 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SH8
 
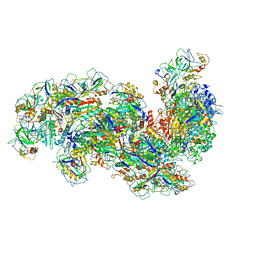 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SHB
 
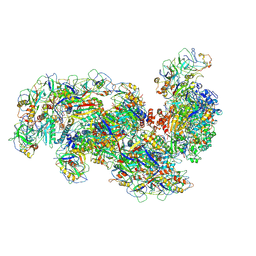 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
1H9H
 
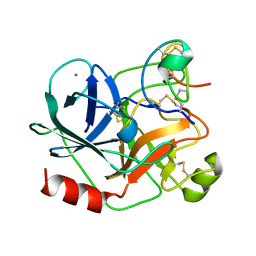 | | COMPLEX OF EETI-II WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
