6PWU
 
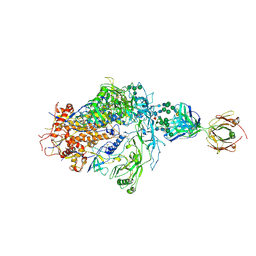 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-07-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6U5O
 
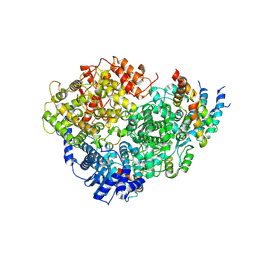 | | Structure of the Human Metapneumovirus Polymerase bound to the phosphoprotein tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Pan, J, Qian, X, Lattmann, S, Sahili, A.E, Yeo, T.H, Kalocsay, M, Fearns, R, Lescar, J. | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the human metapneumovirus polymerase phosphoprotein complex.
Nature, 577, 2020
|
|
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-10-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
3ES5
 
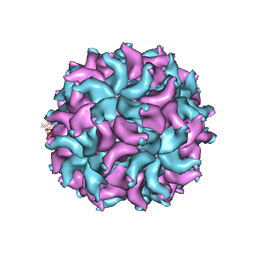 | | Crystal Structure of Partitivirus (PsV-F) | | Descriptor: | Putative capsid protein | | Authors: | Pan, J, Dong, L, Lin, L, Ochoa, W.F, Sinkovits, R.S, Havens, W.M, Nibert, M.L, Baker, T.S, Ghabrial, S.A, Tao, Y.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Atomic structure reveals the unique capsid organization of a dsRNA virus.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2PGG
 
 | |
6SW6
 
 | |
6SW5
 
 | | Crystal structure of the human S-adenosylmethionine synthetase 1 (ligand-free form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, S-adenosylmethionine synthase isoform type-1 | | Authors: | Panmanee, J, Antoyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the dominant inheritance of hypermethioninemia associated with the Arg264His mutation in the MAT1A gene.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3ITM
 
 | | Catalytic domain of hPDE2A | | Descriptor: | ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IBJ
 
 | | X-ray structure of PDE2A | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-07-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ITU
 
 | | hPDE2A catalytic domain complexed with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6G6R
 
 | | Human Methionine Adenosyltransferase II with SAMe and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FCB
 
 | | Human Methionine Adenosyltransferase II mutant (P115G) | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FBP
 
 | | Human Methionine Adenosyltransferase II mutant (S114A) in P22121 crystal form | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FBN
 
 | | Human Methionine Adenosyltransferase II mutant (Q113A) | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FBO
 
 | | Human Methionine Adenosyltransferase II mutant (S114A) in I222 crystal form | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ADENOSINE, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FCD
 
 | | Human Methionine Adenosyltransferase II mutant (R264A) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | Descriptor: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | Authors: | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
8V1T
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1R
 
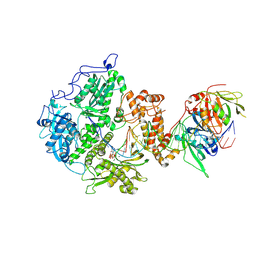 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and DTTP in closed conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1Q
 
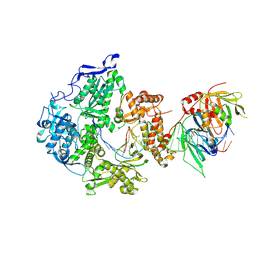 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1S
 
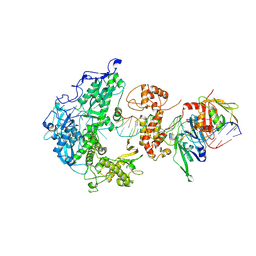 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8U6X
 
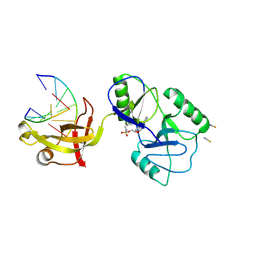 | | ATP-dependent DNA ligase Lig E from Neisseria gonorrhoeae | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*CP*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Williamson, A, Pan, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A role for the ATP-dependent DNA ligase lig E of Neisseria gonorrhoeae in biofilm formation.
Bmc Microbiol., 24, 2024
|
|
7SN0
 
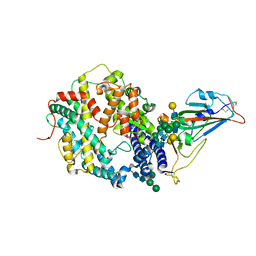 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Clark, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN3
 
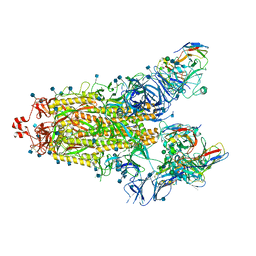 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
 | |
