2LWW
 
 | | NMR structure of RelA-TAD/CBP-TAZ1 complex | | Descriptor: | CREB-binding protein, Nuclear transcription factor RelA, ZINC ION | | Authors: | Mukherjee, S.P, Ghosh, G, Wright, P.E. | | Deposit date: | 2012-08-07 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Analysis of the RelA:CBP/p300 Interaction Reveals Its Involvement in NF-kappa B-Driven Transcription.
Plos Biol., 11, 2013
|
|
2MRE
 
 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2N4K
 
 | | Solution Structure of Enterocin HF, an Antilisterial Bacteriocin Produced by Enterococcus faecium M3K31 | | Descriptor: | Enterocin-HF | | Authors: | Arbulu, S, Lohans, C.T, van Belkum, M.J, Cintas, L.M, Herranz, C, Vederas, J.C, Hernandez, P.E. | | Deposit date: | 2015-06-21 | | Release date: | 2015-12-02 | | Last modified: | 2016-01-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Enterocin HF, an Antilisterial Bacteriocin Produced by Enterococcus faecium M3K31.
J.Agric.Food Chem., 63, 2015
|
|
2MRF
 
 | | NMR structure of the ubiquitin-binding zinc finger (UBZ) domain from human Rad18 | | Descriptor: | E3 ubiquitin-protein ligase RAD18, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2NLA
 
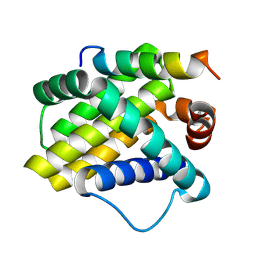 | | Crystal structure of the Mcl-1:mNoxaB BH3 complex | | Descriptor: | FUSION PROTEIN CONSISTING OF Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NL9
 
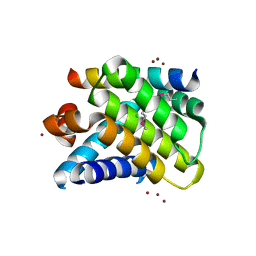 | | Crystal structure of the Mcl-1:Bim BH3 complex | | Descriptor: | Bcl-2-like protein 11, CHLORIDE ION, FUSION PROTEIN CONSISTING OF Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, ... | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NT8
 
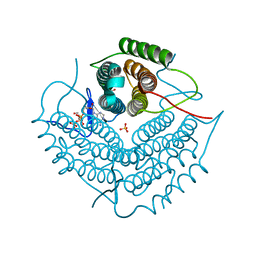 | | ATP bound at the active site of a PduO type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobalamin adenosyltransferase, GLYCEROL, ... | | Authors: | St-Maurice, M, Mera, P.E, Taranto, M.P, Sesma, F, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2006-11-07 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the active site of the PduO-type ATP:Co(I)rrinoid adenosyltransferase from Lactobacillus reuteri.
J.Biol.Chem., 282, 2007
|
|
4BTF
 
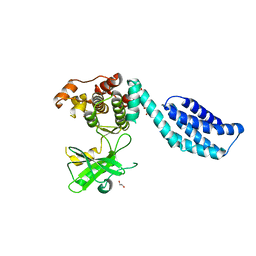 | | Structure of MLKL | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MIXED LINEAGE KINASE DOMAIN-LIKE PROTEIN | | Authors: | Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2013-06-16 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The Pseudokinase Mlkl Mediates Necroptosis Via a Molecular Switch Mechanism
Immunity, 39, 2013
|
|
4C52
 
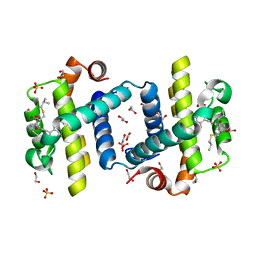 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
4C5D
 
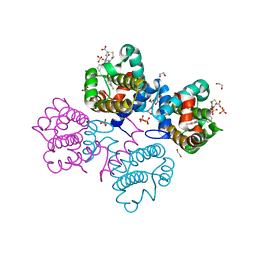 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (42) | | Descriptor: | (R)-3-(4-BROMOBENZYLTHIO)-2-(3-(3-((2,4-DIFLUOROPHENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
4FAS
 
 | |
4F6N
 
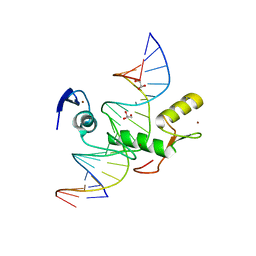 | | Crystal structure of Kaiso zinc finger DNA binding protein in complex with methylated CpG site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*GP*AP*(5CM)P*GP*(5CM)P*GP*GP*TP*GP*AP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*TP*CP*AP*CP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*AP*TP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F6M
 
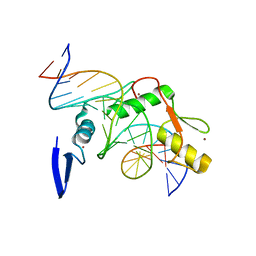 | | Crystal structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
1JVQ
 
 | | Crystal structure at 2.6A of the ternary complex between antithrombin, a P14-P8 reactive loop peptide, and an exogenous tetrapeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2001-08-31 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How small peptides block and reverse serpin polymerisation
J.Mol.Biol., 342, 2004
|
|
1JAW
 
 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1KBH
 
 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
1KDX
 
 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
1L35
 
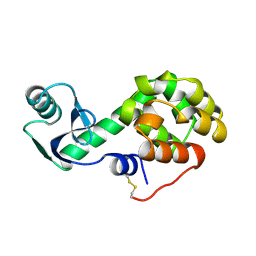 | |
1L8C
 
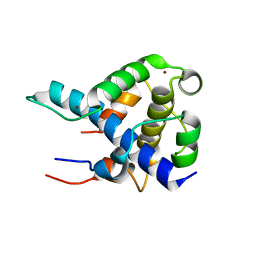 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LK6
 
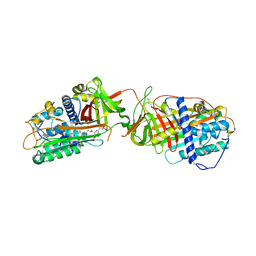 | | Structure of dimeric antithrombin complexed with a P14-P9 reactive loop peptide and an exogenous tripeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2002-04-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serpin Polymerization Is Prevented by a Hydrogen Bond Network That Is Centered on His-334 and Stabilized by Glycerol
J.Biol.Chem., 278, 2003
|
|
1LO1
 
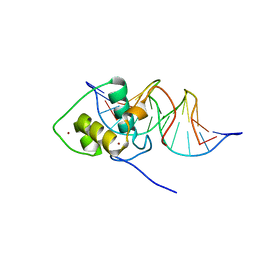 | | ESTROGEN RELATED RECEPTOR 2 DNA BINDING DOMAIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*CP*GP*TP*GP*AP*CP*CP*TP*TP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*AP*AP*GP*GP*TP*CP*AP*CP*G)-3', Steroid hormone receptor ERR2, ... | | Authors: | Gearhart, M.D, Holmbeck, S.M.A, Evans, R.M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-05-05 | | Release date: | 2003-04-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Monomeric Complex of Human Orphan Estrogen Related Receptor-2 with DNA: A Pseudo-dimer Interface Mediates Extended Half-site Recognition
J.Mol.Biol., 327, 2003
|
|
1MAA
 
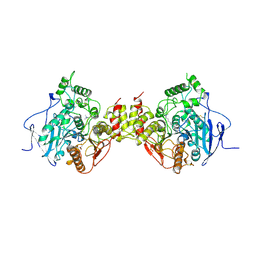 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
1MU8
 
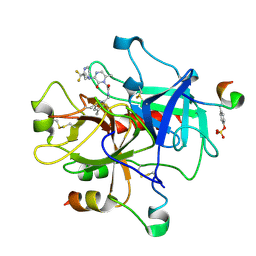 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
1MU6
 
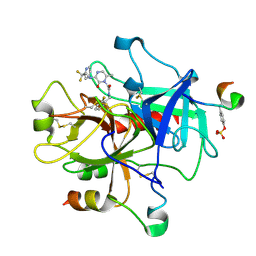 | | Crystal Structure of Thrombin in Complex with L-378,622 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
