6UVG
 
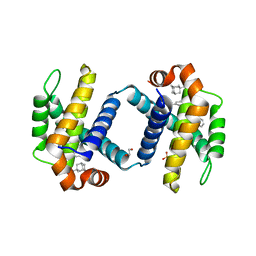 | | Crystal structure of BCL-XL bound to compound 13: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6VUB
 
 | |
6VUF
 
 | |
6VUJ
 
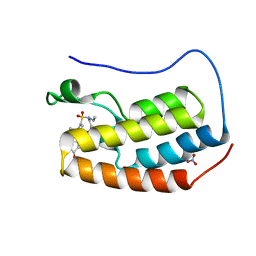 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 15c (N,N-diethyl-3',4'-dimethoxy-6-(1-methyl-5-oxopyrrolidin-3-yl)-[1,1'-biphenyl]-3-sulfonamide) | | Descriptor: | Bromodomain-containing protein 4, N,N-diethyl-3',4'-dimethoxy-6-[(3S)-1-methyl-5-oxopyrrolidin-3-yl][1,1'-biphenyl]-3-sulfonamide, NITRATE ION | | Authors: | Ilyichova, O.V, Scanlon, M.J, Thompson, P.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substituted 1-methyl-4-phenylpyrrolidin-2-ones - Fragment-based design of N-methylpyrrolidone-derived bromodomain inhibitors.
Eur.J.Med.Chem., 191, 2020
|
|
6VUC
 
 | |
2J5D
 
 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
2JPJ
 
 | | Lactococcin G-a in DPC | | Descriptor: | Bacteriocin lactococcin-G subunit alpha | | Authors: | Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P.E. | | Deposit date: | 2007-05-15 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G
Biochim.Biophys.Acta, 1784, 2008
|
|
2JP9
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JOX
 
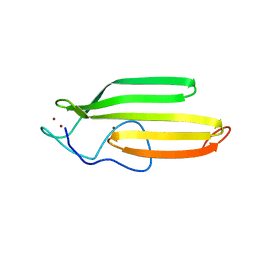 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
2JPA
 
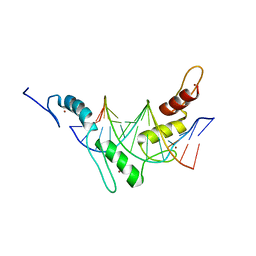 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JM6
 
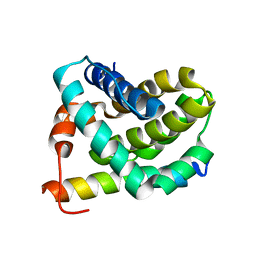 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2JPL
 
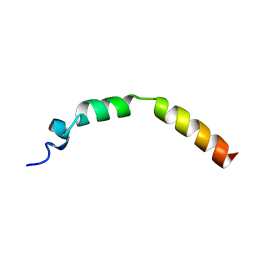 | | Lactococcin G-a in TFE | | Descriptor: | Bacteriocin lactococcin-G subunit alpha | | Authors: | Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P.E. | | Deposit date: | 2007-05-15 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G
Biochim.Biophys.Acta, 1784, 2008
|
|
2JPM
 
 | | Lactococcin G-b in TFE | | Descriptor: | Bacteriocin lactococcin-G subunit beta | | Authors: | Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P.E. | | Deposit date: | 2007-05-15 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G
Biochim.Biophys.Acta, 1784, 2008
|
|
2JPK
 
 | | Lactococcin G-b in DPC | | Descriptor: | Bacteriocin lactococcin-G subunit beta | | Authors: | Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P.E. | | Deposit date: | 2007-05-15 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G
Biochim.Biophys.Acta, 1784, 2008
|
|
2K9Y
 
 | |
2KH2
 
 | | Solution structure of a scFv-IL-1B complex | | Descriptor: | Interleukin-1 beta, scFv | | Authors: | Wilkinson, I.C, Hall, C.J, Veverka, V, Muskett, F.W, Stephens, P.E, Taylor, R.J, Henry, A.J, Carr, M.D. | | Deposit date: | 2009-03-23 | | Release date: | 2009-09-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | High resolution NMR-based model for the structure of a scFv-IL-1beta complex: potential for NMR as a key tool in therapeutic antibody design and development.
J.Biol.Chem., 284, 2009
|
|
2KA4
 
 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2KPE
 
 | | Refined structure of Glycophorin A transmembrane segment dimer in DPC micelles | | Descriptor: | Glycophorin-A | | Authors: | Mineev, K.S, Bocharov, E.V, Goncharuk, M.V, Arseniev, A.S, Volynsky, P.E, Efremov, R.G. | | Deposit date: | 2009-10-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments.
Acta Naturae, 3, 2011
|
|
2KHF
 
 | |
2KA6
 
 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
2KJE
 
 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | Descriptor: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | Authors: | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KPF
 
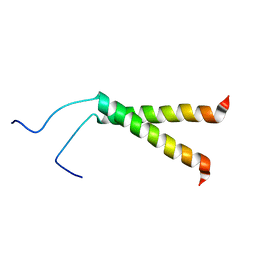 | | Spatial structure of the dimeric transmembrane domain of glycophorin A in bicelles soluton | | Descriptor: | Glycophorin-A | | Authors: | Mineev, K.S, Bocharov, E.V, Goncharuk, M.V, Arseniev, A.S, Volynsky, P.E, Efremov, R.G. | | Deposit date: | 2009-10-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments.
Acta Naturae, 3, 2011
|
|
2L14
 
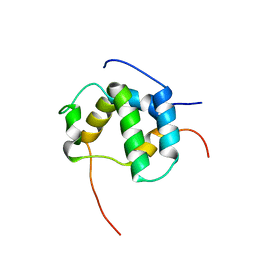 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2LT7
 
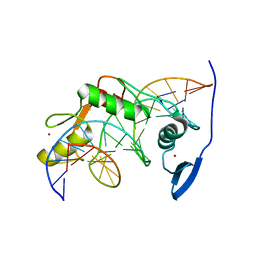 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LEF
 
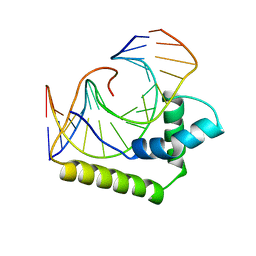 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
