2RPT
 
 | |
6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | Descriptor: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | Authors: | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
2V7F
 
 | | Structure of P. abyssi RPS19 protein | | Descriptor: | CHLORIDE ION, RPS19E SSU RIBOSOMAL PROTEIN S19E | | Authors: | Gregory, L.A, Aguissa-Toure, A.H, Pinaud, N, Legrand, P, Gleizes, P.E, Fribourg, S. | | Deposit date: | 2007-07-30 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Basis of Diamond Blackfan Anemia: Structure and Function Analysis of Rps19.
Nucleic Acids Res., 35, 2007
|
|
2V94
 
 | |
2VOF
 
 | | Structure of mouse A1 bound to the Puma BH3-domain | | Descriptor: | BCL-2-BINDING COMPONENT 3, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VTY
 
 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
2VOG
 
 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOI
 
 | | Structure of mouse A1 bound to the Bid BH3-domain | | Descriptor: | BCL-2-RELATED PROTEIN A1, BH3-INTERACTING DOMAIN DEATH AGONIST P13, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOH
 
 | | Structure of mouse A1 bound to the Bak BH3-domain | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-2-RELATED PROTEIN A1, CITRIC ACID, ... | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
4Z6I
 
 | | Crystal structure of BRD9 bromodomain in complex with a substituted valerolactam quinolone ligand | | Descriptor: | Bromodomain-containing protein 9, tert-butyl [(2R,3S)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxo-2-phenylpiperidin-3-yl]carbamate | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ZIE
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Krishna Kumar, K, Robin, A.Y, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIH
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZQL
 
 | | Crystal structure of TRIM24 with 3,4-dimethoxy-N-(6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)benzenesulfonamide inhibitor | | Descriptor: | 3,4-dimethoxy-N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, DIMETHYL SULFOXIDE, PENTAETHYLENE GLYCOL, ... | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of TRIM24 with 3,4-dimethoxy-N-(6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)benzenesulfonamide inhibitor
To Be Published
|
|
4Z6H
 
 | | Crystal structure of BRD9 bromodomain in complex with a valerolactam quinolone ligand | | Descriptor: | 1,4-dimethyl-7-(2-oxopiperidin-1-yl)quinolin-2(1H)-one, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ZIF
 
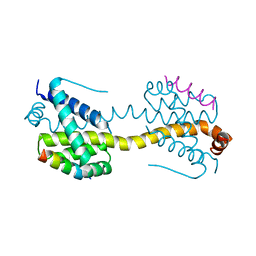 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZL4
 
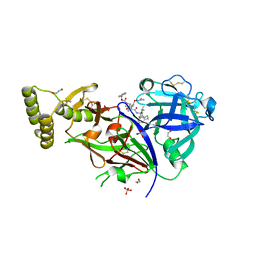 | | Plasmepsin V from Plasmodium vivax bound to a transition state mimetic (WEHI-842) | | Descriptor: | (2R)-1-[(2R)-2-(2-methoxyethoxy)propoxy]propan-2-amine, 1,2-ETHANEDIOL, Aspartic protease PM5, ... | | Authors: | Czabotar, P.E, Hodder, A.N, Smith, B.J, Sleebs, B.E, Gazdic, M, Boddey, J.A, Cowman, A.F. | | Deposit date: | 2015-05-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for plasmepsin V inhibition that blocks export of malaria proteins to human erythrocytes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4ZII
 
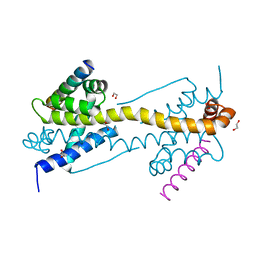 | | Crystal Structure of core/latch dimer of BaxI66A in complex with BidBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Czabotar, P.E, Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIG
 
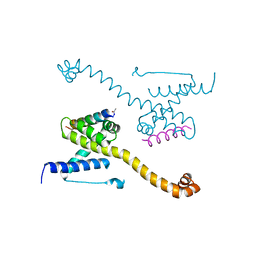 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
5A2F
 
 | | Two membrane distal IgSF domains of CD166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD166 ANTIGEN | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
5A2E
 
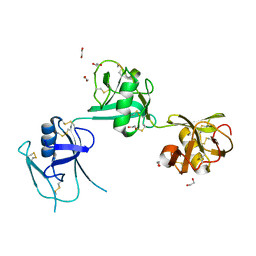 | | Extracellular SRCR domains of human CD6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-CELL DIFFERENTIATION ANTIGEN CD6 | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
5ACZ
 
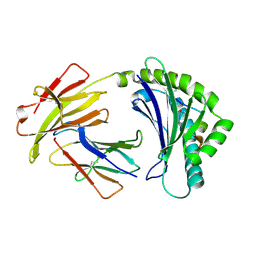 | | COMPLEX OF A B21 CHICKEN MHC CLASS I MOLECULE AND A 11MER CHICKEN PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 11MER PEPTIDE, BETA-2-MICROGLOBULIN, ... | | Authors: | Chappell, P.E, Roversi, P, Harrison, M.C, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Complex of a B21 Chicken Mhc Class I Molecule and a 11mer Chicken Peptide
To be Published
|
|
5AD0
 
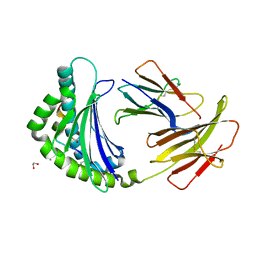 | | COMPLEX OF A B21 CHICKEN MHC CLASS I MOLECULE AND A 11MER CHICKEN PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 11MER PEPTIDE, BETA-2-MICROGLOBULIN, ... | | Authors: | Chappell, P.E, Roversi, P, Harrison, M.C, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Complex of a B21 Chicken Mhc Class I Molecule and a 11mer Chicken Peptide
To be Published
|
|
5AQ6
 
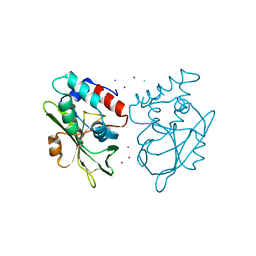 | | Structure of E. coli ZinT at 1.79 Angstrom | | Descriptor: | ACETIC ACID, METAL-BINDING PROTEIN ZINT, ZINC ION | | Authors: | Santo, P.E, Colaco, H.G, Matias, P, Vicente, J.B, Bandeiras, T.M. | | Deposit date: | 2015-09-21 | | Release date: | 2016-01-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Roles of Escherichia Coli Zint in Cobalt, Mercury and Cadmium Resistance and Structural Insights Into the Metal Binding Mechanism
Metallomics, 8, 2016
|
|
5BT4
 
 | | Crystal structure of BRD4 first bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of BRD4 first bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BT5
 
 | | Crystal structure of BRD2 second bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BRD2 second bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
