9JBQ
 
 | | Structure of the complex between h1F3 Fab and PcrV fragment | | Descriptor: | Heavy chain, PcrV, light chain | | Authors: | Numata, S, Hara, T, Izawa, M, Okuno, Y, Sato, Y, Yamane, S, Maki, H, Sato, T, Yamano, Y. | | Deposit date: | 2024-08-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel humanized anti-PcrV monoclonal antibody COT-143 protects mice from lethal Pseudomonas aeruginosa infection via inhibition of toxin translocation by the type III secretion system.
Antimicrob.Agents Chemother., 68, 2024
|
|
6KSL
 
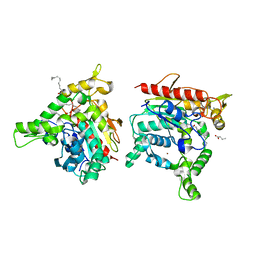 | | Staphylococcus aureus lipase - S116A inactive mutant | | Descriptor: | CALCIUM ION, LAURIC ACID, Lipase 2, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
8H7X
 
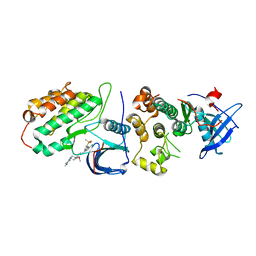 | | Crystal structure of EGFR T790M/C797S mutant in complex with brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
NPJ Precis Oncol, 8, 2024
|
|
6FEK
 
 | |
4WUA
 
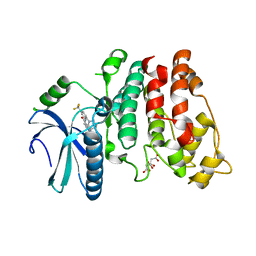 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | Descriptor: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | Authors: | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
3WHE
 
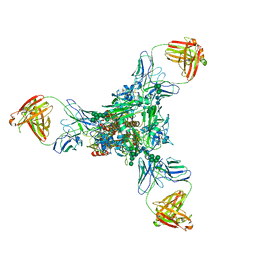 | | A new conserved neutralizing epitope at the globular head of hemagglutinin in H3N2 influenza viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fujii, Y, Sumida, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-08-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses.
J.Virol., 88, 2014
|
|
5C0R
 
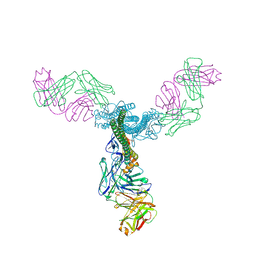 | | Crystal Structure of a Generation 3 Influenza Hemagglutinin Stabilized Stem Complexed with the Broadly Neutralizing Antibody C179 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C179 Fab heavy chain, ... | | Authors: | Boyington, J.C, kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
5C0S
 
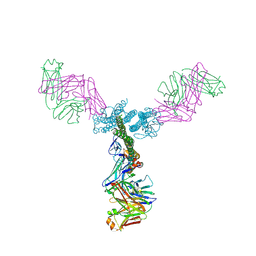 | | Crystal structure of a generation 4 influenza hemagglutinin stabilized stem in complex with the broadly neutralizing antibody CR6261 | | Descriptor: | CR6261 antibody heavy chain, CR6261 antibody light chain, Hemagglutinin, ... | | Authors: | Boyington, J.C, Kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
4O58
 
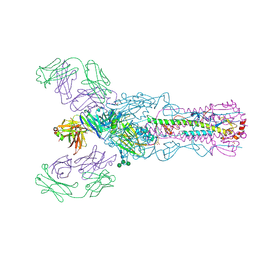 | |
4O5N
 
 | | Crystal structure of A/Victoria/361/2011 (H3N2) influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
4O5L
 
 | | Crystal structure of broadly neutralizing antibody F045-092 | | Descriptor: | Fab F045-092 heavy chain, Fab F045-092 light chain, PHOSPHATE ION | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5045 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
4O5I
 
 | |
7DKJ
 
 | |
8YIB
 
 | | Staphylococcus aureus lipase -PSA complex - covalent bonding state | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-02-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Staphylococcus aureus lipase complex with unsaturated petroselinic acid.
Febs Open Bio, 14, 2024
|
|
8K7Q
 
 | | Staphylococcus aureus lipase S116A inactive mutant-PSA complex | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Staphylococcus aureus lipase complex with unsaturated petroselinic acid.
Febs Open Bio, 14, 2024
|
|
8K7P
 
 | | Staphylococcus aureus lipase -PSA complex | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Staphylococcus aureus lipase complex with unsaturated petroselinic acid.
Febs Open Bio, 14, 2024
|
|
6KSM
 
 | | Staphylococcus aureus lipase -Orlistat complex | | Descriptor: | (2S,3S,5S)-5-[(N-FORMYL-L-LEUCYL)OXY]-2-HEXYL-3-HYDROXYHEXADECANOIC ACID, CALCIUM ION, LAURIC ACID, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
6KSI
 
 | | Staphylococcus aureus lipase - native | | Descriptor: | CALCIUM ION, HEXANOIC ACID, LAURIC ACID, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
7DPJ
 
 | | H-Ras Q61L in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Taniguchi, H, Matsumoto, S, Miyamoto, R, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
1MPG
 
 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
