1WAQ
 
 | |
4AJV
 
 | | Structure of mouse ZP-C domain of TGF-Beta-Receptor-3 | | Descriptor: | TRANSFORMING GROWTH FACTOR BETA RECEPTOR TYPE 3 | | Authors: | Diestel, U, Resch, M, Meinhardt, K, Weiler, S, Hellmann, T.V, Nickel, J, Eichler, J, Muller, Y.A. | | Deposit date: | 2012-02-20 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a Novel Tgf-Beta-Binding Site in the Zona Pellucida C-Terminal (Zp-C) Domain of Tgf-Beta-Receptor-3 (Tgfr-3).
Plos One, 8, 2013
|
|
1REU
 
 | | Structure of the bone morphogenetic protein 2 mutant L51P | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, bone morphogenetic protein 2 | | Authors: | Keller, S, Nickel, J, Zhang, J.-L, Sebald, W, Mueller, T.D. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular recognition of BMP-2 and BMP receptor IA.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1REW
 
 | | Structural refinement of the complex of bone morphogenetic protein 2 and its type IA receptor | | Descriptor: | bone morphogenetic protein 2, bone morphogenetic protein receptor type IA | | Authors: | Keller, S, Nickel, J, Zhang, J.-L, Sebald, W, Mueller, T.D. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Molecular recognition of BMP-2 and BMP receptor IA.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3QB4
 
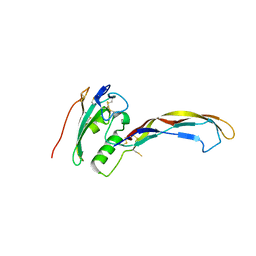 | |
3QT2
 
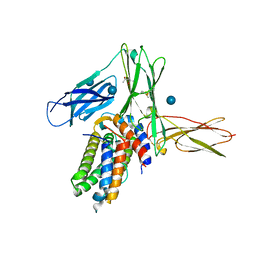 | | Structure of a cytokine ligand-receptor complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Interleukin-5, Interleukin-5 receptor subunit alpha, ... | | Authors: | Mueller, T.D, Patino, E, Kotzsch, A, Saremba, S, Nickel, J, Schmitz, W, Sebald, W. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure Analysis of the IL-5 Ligand-Receptor Complex Reveals a Wrench-like Architecture for IL-5Ralpha.
Structure, 19, 2011
|
|
3EVS
 
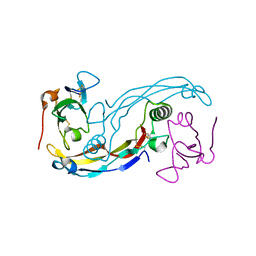 | | Crystal structure of the GDF-5:BMP receptor IB complex. | | Descriptor: | Bone morphogenetic protein receptor type-1B, Growth/differentiation factor 5 | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity.
Embo J., 28, 2009
|
|
2R52
 
 | |
2R53
 
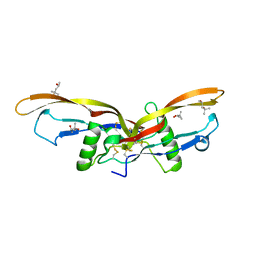 | |
2K3G
 
 | |
2QJA
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant B12 | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
2QJB
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant IA/IB | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA, CHLORIDE ION | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
2QJ9
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant B1 | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
2H64
 
 | |
2H62
 
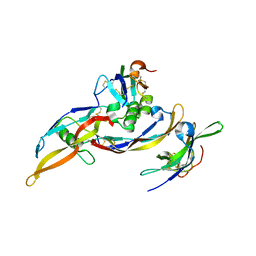 | | Crystal structure of a ternary ligand-receptor complex of BMP-2 | | Descriptor: | Acvr2b protein, Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Mueller, T.D. | | Deposit date: | 2006-05-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor.
Bmc Struct.Biol., 7, 2007
|
|
6H41
 
 | |
2B8X
 
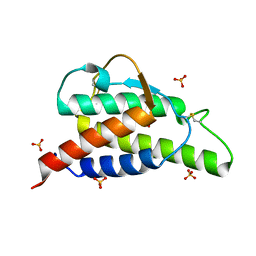 | | Crystal structure of the interleukin-4 variant F82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Z
 
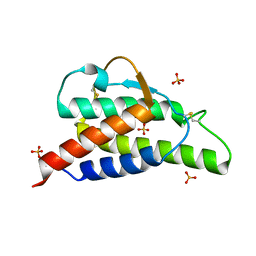 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2D48
 
 | | Crystal structure of the Interleukin-4 variant T13D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B90
 
 | | Crystal structure of the interleukin-4 variant T13DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8U
 
 | | Crystal structure of wildtype human Interleukin-4 | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Y
 
 | | Crystal structure of the interleukin-4 variant T13DF82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B91
 
 | | Crystal structure of the interleukin-4 variant F82DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
1ES7
 
 | |
2MQH
 
 | |
