4HOQ
 
 | | Crystal Structure of Full-Length Human IFIT5 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5 | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
6DLU
 
 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
8EEW
 
 | | CryoEM of the soluble OPA1 dimer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFF
 
 | | CryoEM of the soluble OPA1 tetramer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFS
 
 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EF7
 
 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFT
 
 | | CryoEM of the soluble OPA1 interfaces from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFR
 
 | | CryoEM of the soluble OPA1 interfaces with GDP-AlFx bound from the helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
1SEK
 
 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
6DLV
 
 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
2H8L
 
 | |
6MHM
 
 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dementiev, A, Joachimiak, A, Doan, N. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
1QSJ
 
 | | N-TERMINALLY TRUNCATED C3DG FRAGMENT | | Descriptor: | COMPLEMENT C3 PRECURSOR | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Folli, C, Berni, R. | | Deposit date: | 1999-06-22 | | Release date: | 2000-07-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
4K1O
 
 | |
4K1N
 
 | |
4K7D
 
 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
1QQF
 
 | | N-TERMINALLY TRUNCATED C3D,G FRAGMENT OF THE COMPLEMENT SYSTEM | | Descriptor: | PROTEIN (COMPLEMENT C3DG) | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Berni, R. | | Deposit date: | 1999-06-04 | | Release date: | 2000-07-31 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
3QLG
 
 | |
3QLF
 
 | |
3OEZ
 
 | | crystal structure of the L317I mutant of the chicken c-Src tyrosine kinase domain complexed with imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Boubeva, R, Pernot, L, Perozzo, R, Scapozza, L. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | a single amino-acid dictates the dynamics of the switch between active and inactive C-Src conformation
To be Published
|
|
3OF0
 
 | |
4ERK
 
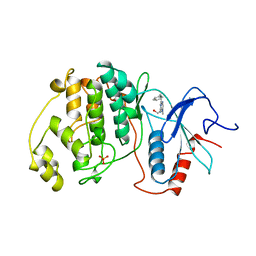 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/OLOMOUCINE | | Descriptor: | EXTRACELLULAR REGULATED KINASE 2, OLOMOUCINE, SULFATE ION | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
8T0R
 
 | |
8SZ8
 
 | |
8SZ7
 
 | |
