1EWY
 
 | | ANABAENA PCC7119 FERREDOXIN:FERREDOXIN-NADP+-REDUCTASE COMPLEX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I, FERREDOXIN-NADP REDUCTASE, ... | | Authors: | Morales, R, Charon, M.H, Frey, M. | | Deposit date: | 2000-04-28 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic studies of the interaction between the ferredoxin-NADP+ reductase and ferredoxin from the cyanobacterium Anabaena: looking for the elusive ferredoxin molecule.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QT9
 
 | | OXIDIZED [2FE-2S] FERREDOXIN FROM ANABAENA PCC7119 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Morales, R, Chron, M.-H, Hudry-Clergeon, G, Petillot, Y, Norager, S, Medina, M, Frey, M. | | Deposit date: | 1999-07-01 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined X-ray structures of the oxidized, at 1.3 A, and reduced, at 1.17 A, [2Fe-2S] ferredoxin from the cyanobacterium Anabaena PCC7119 show redox-linked conformational changes.
Biochemistry, 38, 1999
|
|
3VCM
 
 | | Crystal structure of human prorenin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, prorenin | | Authors: | Morales, R, Watier, Y, Bocskei, Z. | | Deposit date: | 2012-01-04 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Human Prorenin Structure Sheds Light on a Novel Mechanism of Its Autoinhibition and on Its Non-Proteolytic Activation by the (Pro)renin Receptor.
J.Mol.Biol., 421, 2012
|
|
1CZP
 
 | | ANABAENA PCC7119 [2FE-2S] FERREDOXIN IN THE REDUCED AND OXIXIZED STATE AT 1.17 A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Morales, R, Charon, M.H, Frey, M. | | Deposit date: | 1999-09-06 | | Release date: | 2000-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Refined X-ray structures of the oxidized, at 1.3 A, and reduced, at 1.17 A, [2Fe-2S] ferredoxin from the cyanobacterium Anabaena PCC7119 show redox-linked conformational changes.
Biochemistry, 38, 1999
|
|
1ROS
 
 | | Crystal structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-[2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL)ETHYL]-4-(4'-ETHOXY-1,1'-BIPHENYL-4-YL)-4-OXOBUTANOIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J.Mol.Biol., 341, 2004
|
|
1UTZ
 
 | | Crystal Structure of MMP-12 complexed to (2R)-3-({[4-[(pyridin-4-yl)phenyl]-thien-2-yl}carboxamido)(phenyl)propanoic acid | | Descriptor: | (2R)-3-({[4-[(PYRIDIN-4-YL)PHENYL]-THIEN-2-YL}CARBOXAMIDO)(PHENYL)PROPANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Novel Non-Peptidic, Non-Zinc Chelating Inhibitors Bound to Mmp-12
J.Mol.Biol., 341, 2004
|
|
1UTT
 
 | | Crystal Structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL) ETHYL-4-(4'-ETHOXY [1,1'-BIPHENYL]-4-YL)-4-OXOBUTANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J. Mol. Biol., 341, 2004
|
|
2V3Q
 
 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
4G11
 
 | |
4W4W
 
 | | JNK2/3 in complex with N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide | | Descriptor: | N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4V
 
 | | JNK2/3 in complex with 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4Y
 
 | | JNK2/3 in complex with 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4X
 
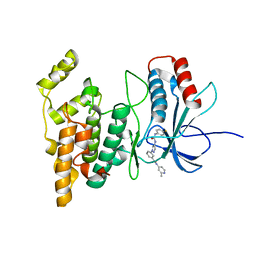 | | JNK2/3 in complex with 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
5KKG
 
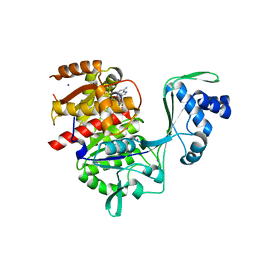 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
5K27
 
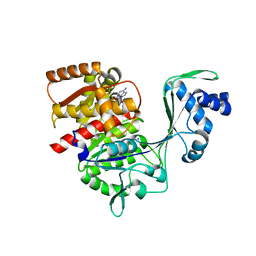 | | Crystal structure of ancestral protein ancMT of ADP-dependent sugar kinases family. | | Descriptor: | ADENOSINE MONOPHOSPHATE, IODIDE ION, ancMT | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1W5T
 
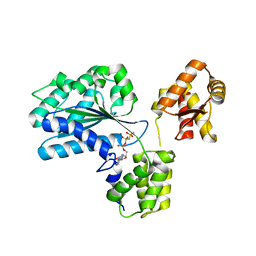 | | Structure of the Aeropyrum Pernix ORC2 protein (ADPNP-ADP complexes) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ORC2, ... | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
1W5S
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADP form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ORIGIN RECOGNITION COMPLEX SUBUNIT 2 ORC2, SULFATE ION | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
5FUQ
 
 | | CRYSTAL STRUCTURE OF THE H80R VARIANT OF NQO1 BOUND TO DICOUMAROL | | Descriptor: | ACETATE ION, BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gavira, J.A, Medina-Carmona, E, Pey, A.L. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Enhanced vulnerability of human proteins towards disease-associated inactivation through divergent evolution.
Hum.Mol.Genet., 26, 2017
|
|
6TDM
 
 | | Bam_5920cDD 5919nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDN
 
 | | Bam_5925cDD 5924nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDD
 
 | | Bam_5924 docking domain | | Descriptor: | Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6HGP
 
 | |
6HGS
 
 | | Crystal Structure of Human APRT wild type in complex with GMP | | Descriptor: | Adenine phosphoribosyltransferase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for substrate selectivity and nucleophilic substitution mechanisms in human adenine phosphoribosyltransferase catalyzed reaction.
J.Biol.Chem., 294, 2019
|
|
6HGR
 
 | | Crystal Structure of Human APRT wild type in complex with IMP | | Descriptor: | Adenine phosphoribosyltransferase, INOSINIC ACID | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis for substrate selectivity and nucleophilic substitution mechanisms in human adenine phosphoribosyltransferase catalyzed reaction.
J.Biol.Chem., 294, 2019
|
|
