1N1L
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND INHIBITOR (GW472467X) | | Descriptor: | HCV NS3 SERINE PROTEASE, NS4A COFACTOR, ZINC ION, ... | | Authors: | Andrews, D.M, Chaignot, H, Coomber, B.A, Good, A.C, Hind, S.L, Jones, P.S, Mill, G, Robinson, J.E, Skarzynski, T, Slater, M.J, Somers, D.O.N. | | Deposit date: | 2002-10-18 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 2. The use of X-ray Crystal Structure Data in the Optimisation of P3 and P4 Substituents
Org.Lett., 4, 2002
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
8T3V
 
 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | Descriptor: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3O
 
 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3Q
 
 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3S
 
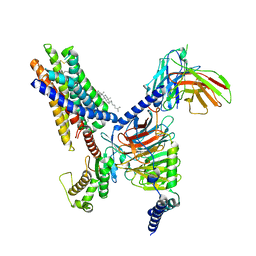 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | Descriptor: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
6H0K
 
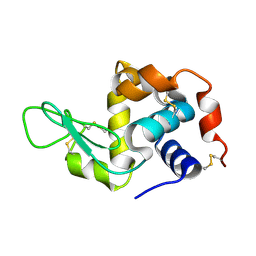 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
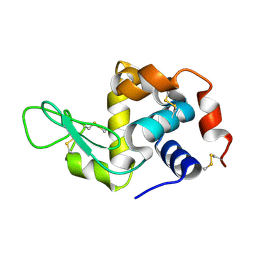 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6PFY
 
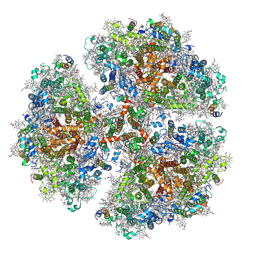 | |
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
8BEY
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 7 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEX
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 3 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEZ
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 11 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7K8E
 
 | | Beta-lactamase mixed with Ceftriaxone, 5ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40005636 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8F
 
 | | Beta-lactamase mixed with Ceftriaxone, 10ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60003138 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8H
 
 | | Beta-lactamase mixed with Ceftriaxone, 50ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60006261 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8K
 
 | | Beta-lactamase mixed with Sulbactam, 60ms | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULBACTAM, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8L
 
 | | Beta-lactamase, Unmixed | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8000102 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
1RTL
 
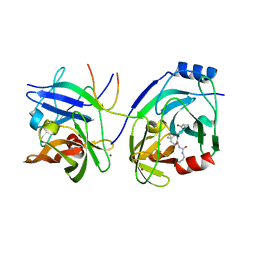 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN: NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND PYRROLIDINE-5,5-TRANSLACTAM INHIBITOR | | Descriptor: | N-[(2R,3S)-1-((2S)-2-{[(CYCLOPENTYLAMINO)CARBONYL]AMINO}-3-METHYLBUTANOYL)-2-(1-FORMYL-1-CYCLOBUTYL)PYRROLIDINYL]CYCLOPROPANECARBOXAMIDE, NS3 protease/helicase, NS4A COFACTOR, ... | | Authors: | Skarzynski, T, Somers, D.O.N. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease
Org.Lett., 5, 2003
|
|
6GW9
 
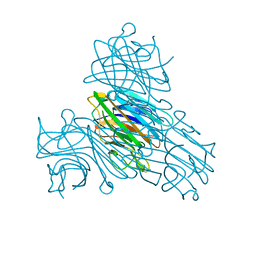 | | Concanavalin A structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | CALCIUM ION, Concanavalin V, MAGNESIUM ION | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6GWA
 
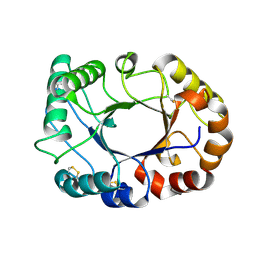 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6PGK
 
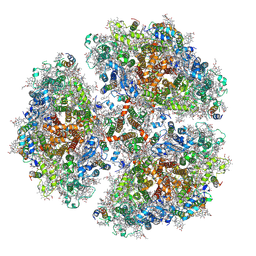 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Gisriel, C, Fromme, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
7TUM
 
 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
