3ZZM
 
 | | Crystal structure of Mycobacterium tuberculosis PurH with a novel bound nucleotide CFAIR, at 2.2 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, GLYCEROL, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-02 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
4A1O
 
 | | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
8G8P
 
 | |
3KLQ
 
 | | Crystal Structure of the Minor Pilin FctB from Streptococcus pyogenes 90/306S | | Descriptor: | GLYCEROL, Putative pilus anchoring protein | | Authors: | Linke, C, Young, P.G, Bunker, R.D, Caradoc-Davies, T.T, Baker, E.N. | | Deposit date: | 2009-11-08 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring
J.Biol.Chem., 285, 2010
|
|
4PMK
 
 | |
6XPP
 
 | |
5U6F
 
 | |
5U5O
 
 | |
4C0Z
 
 | | The N-terminal domain of the Streptococcus pyogenes pilus tip adhesin Cpa | | Descriptor: | ANCILLARY PROTEIN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Linke-Winnebeck, C, Paterson, N, Baker, E.N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-11-20 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Model for the Covalent Adhesion of the Streptococcus Pyogenes Pilus Through a Thioester Bond.
J.Biol.Chem., 289, 2014
|
|
7ULE
 
 | | F420-1/GDP complex of F420-gamma glutamyl ligase (CofE) from Archaeoglobus fulgidus | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[oxidanyl-[(2~{R},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-[2,4,8-tris(oxidanylidene)-1,9-dihydropyrimido[4,5-b]quinolin-10-yl]pentoxy]phosphoryl]oxypropanoyl]amino]pentanedioic acid, Coenzyme F420:L-glutamate ligase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bashiri, G, Squire, C.J. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Poly-gamma-glutamylation of biomolecules.
Nat Commun, 15, 2024
|
|
7ULD
 
 | |
7ULF
 
 | |
6MZQ
 
 | | TAS-120 in reversible binding mode with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1 | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
6MZW
 
 | | TAS-120 covalent complex with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
6NVJ
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVL
 
 | | FGFR1 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 1, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVG
 
 | | FGFR4 complex with N-(3,5-dichloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)phenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3,5-dichloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)phenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVK
 
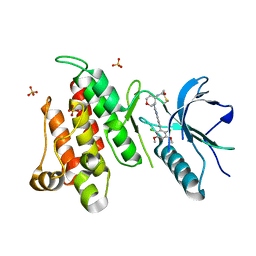 | | FGFR4 complex with BLU-554, N-((3S,4S)-3-((6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl)amino)tetrahydro-2H-pyran-4-yl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3S,4S)-3-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}oxan-4-yl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVI
 
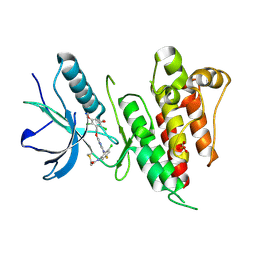 | | FGFR4 complex with N-(3-chloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-5-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3-chloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-5-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVH
 
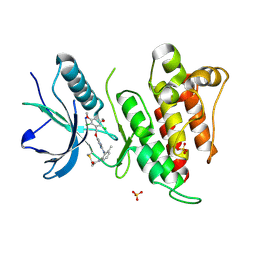 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
