5TR1
 
 | |
5U70
 
 | |
5VA3
 
 | |
5VA1
 
 | |
8DDX
 
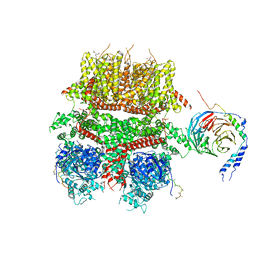 | | cryo-EM structure of TRPM3 ion channel in complex with Gbg in the presence of PIP2, tethered by ALFA-nanobody | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
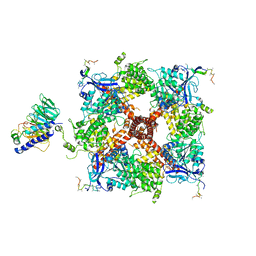
8DDW
 
 | |
8DDR
 
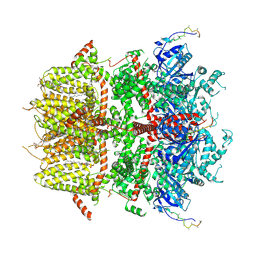 | | cryo-EM structure of TRPM3 ion channel in the absence of PIP2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDT
 
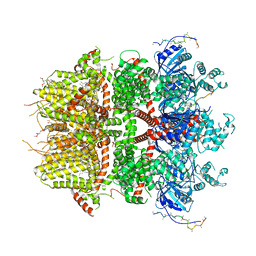 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDS
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDV
 
 | | Cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDU
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED7
 
 | | cryo-EM structure of TRPM3 ion channel in apo state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Transient receptor potential cation channel, subfamily M, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED8
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2 and PregS, state 1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED9
 
 | | cryo-EM structure of TRPM3 ion channel in the presence with PIP2 and PregS, state 2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8EMW
 
 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on liposomes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EMX
 
 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on lipid nanodiscs | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EMV
 
 | | Phospholipase C beta 3 (PLCb3) in solution | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3SPI
 
 | | Inward rectifier potassium channel Kir2.2 in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPH
 
 | | Inward rectifier potassium channel Kir2.2 I223L mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SYA
 
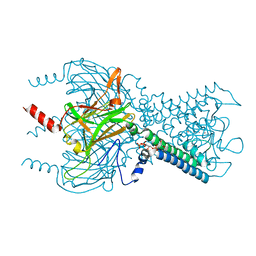 | | Crystal structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) in complex with sodium and PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Whorton, M.R, MacKinnon, R. | | Deposit date: | 2011-07-16 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal Structure of the Mammalian GIRK2 K(+) Channel and Gating Regulation by G Proteins, PIP(2), and Sodium.
Cell(Cambridge,Mass.), 147, 2011
|
|
3SPC
 
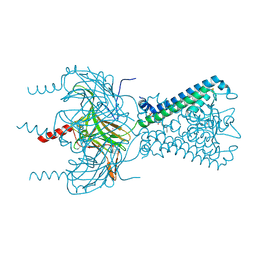 | | Inward rectifier potassium channel Kir2.2 in complex with dioctanoylglycerol pyrophosphate (DGPP) | | Descriptor: | (2R)-3-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}propane-1,2-diyl dioctanoate, Inward-rectifier K+ channel Kir2.2, POTASSIUM ION | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SYC
 
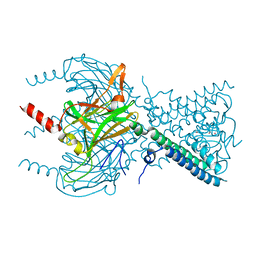 | |
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SYP
 
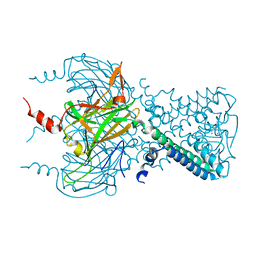 | |
