5HRM
 
 | |
5IOJ
 
 | |
3LL5
 
 | |
3LKK
 
 | |
7UVR
 
 | | Crystal structure of human ClpP protease in complex with TR-65 | | Descriptor: | 3-{[(10R)-4-[(4-chlorophenyl)methyl]-5-oxo-1,2,4,5,8,9-hexahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-7(6H)-yl]methyl}benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UW0
 
 | | Crystal structure of human ClpP protease in complex with TR-133 | | Descriptor: | 3-({3-[(4-bromophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVM
 
 | | Crystal structure of human ClpP protease in complex with TR-27 | | Descriptor: | (10R)-4-[(4-chlorophenyl)methyl]-7-[(3-ethynylphenyl)methyl]-2,4,6,7,8,9-hexahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(1H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVN
 
 | | Crystal structure of human ClpP protease in complex with TR-57 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-1-methyl-2,4-dioxo-1,3,4,5,7,8-hexahydropyrido[4,3-d]pyrimidin-6(2H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVU
 
 | | Crystal structure of human ClpP protease in complex with TR-107 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
6WN6
 
 | | Crystal structure of 3-keto-D-glucoside 4-epimerase, YcjR, from E. coli, apo form | | Descriptor: | 1,2-ETHANEDIOL, 3-keto-D-glucoside 4-epimerase, MANGANESE (II) ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Mukherjee, K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Reaction Mechanism of YcjR, an Epimerase That Facilitates the Interconversion of d-Gulosides to d-Glucosides inEscherichia coli.
Biochemistry, 59, 2020
|
|
9B9H
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40333 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-({(17E)-18-[(3P)-4-{[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-16-oxo-3,6,9,12-tetraoxa-15-azaoctadec-17-en-1-yl}carbamoyl)-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Alvarez, H.G, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-02 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9B9W
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40792 | | Descriptor: | (4P)-N-[(1R)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-[(25E)-1-{(1P)-6-fluoro-3'-[4-fluoro-2-(trifluoromethyl)benzamido]-4'-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-1,24-dioxo-5,8,11,14,17,20-hexaoxa-2,23-diazahexacos-25-en-26-yl]-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Mamai, A, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9BA2
 
 | | Crystal structure of the binary complex of DCAF1 and WDR5 | | Descriptor: | DDB1- and CUL4-associated factor 1, IMIDAZOLE, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Srivastava, S, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9B9T
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40407 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-[(17-{(4P)-4-[(2P)-4-{[(1R)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-1H-pyrazol-1-yl}-16-oxo-3,6,9,12-tetraoxa-15-azaheptadecan-1-yl)carbamoyl]-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Krausser, C, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9DLW
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-41114 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-({(32E)-33-[(3P)-4-{[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-31-oxo-3,6,9,12,15,18,21,24,27-nonaoxa-30-azatritriacont-32-en-1-yl}carbamoyl)-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Mamai, A, Wilson, B.J, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-09-11 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
4L9P
 
 | | Crystal structure of Aspergillus fumigatus protein farnesyltransferase complexed with the FII analog, FPT-II, and the KCVVM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CaaX farnesyltransferase alpha subunit Ram2, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-06-18 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
4LNG
 
 | | Aspergillus fumigatus protein farnesyltransferase complex with farnesyldiphosphate and tipifarnib | | Descriptor: | 1,2-ETHANEDIOL, 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, CaaX farnesyltransferase alpha subunit Ram2, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
4MBG
 
 | | Crystal structure of Aspergillus fumigatus protein farnesyltransferase binary complex with farnesyldiphosphate | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
4LNB
 
 | | Aspergillus fumigatus protein farnesyltransferase ternary complex with farnesyldiphosphate and ethylenediamine scaffold inhibitor 5 | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
6NAH
 
 | |
6NAW
 
 | |
6NB1
 
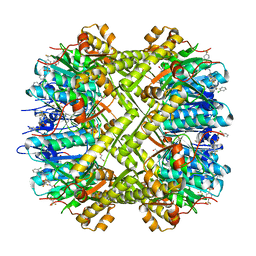 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
6BBA
 
 | |
8SZM
 
 | |
