3JSZ
 
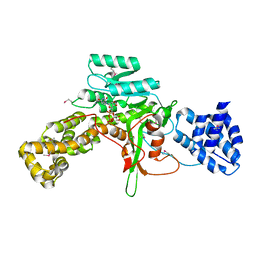 | | Legionella pneumophila glucosyltransferase Lgt1 N293A with UDP-Glc | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | Deposit date: | 2009-09-11 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
3JT1
 
 | | Legionella pneumophila glucosyltransferase Lgt1, UDP-bound form | | Descriptor: | Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | Deposit date: | 2009-09-11 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
5UP2
 
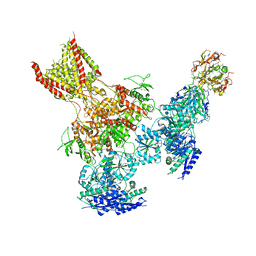 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, Ro 25-6981, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GluN2B-specific Fab, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
5UOW
 
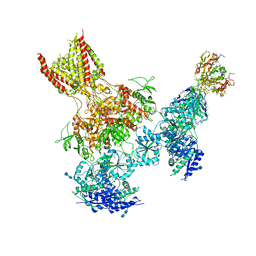 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
8XFS
 
 | | LGR4-RSPO2-ZNRF3 RING domain (1:2:2) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, Leucine-rich repeat-containing G-protein coupled receptor 4, R-spondin-2, ... | | Authors: | Lu, W, Yong, G. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | LGR4-RSPO2-ZNRF3 RING domain (1:2:2)
To Be Published
|
|
8XFT
 
 | | LGR4-RSPO2-ZNRF3(1:1:1) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, Leucine-rich repeat-containing G-protein coupled receptor 4, MB52, ... | | Authors: | Lu, W, Yong, G. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | LGR4-RSPO2-ZNRF3(1:1:1)
To Be Published
|
|
6UIW
 
 | |
6UIV
 
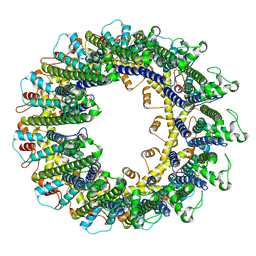 | |
6UIX
 
 | | Cryo-EM structure of human CALHM2 gap junction | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Lu, W, Du, J, Choi, W. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structures and gating mechanism of human calcium homeostasis modulator 2.
Nature, 576, 2019
|
|
8HZ8
 
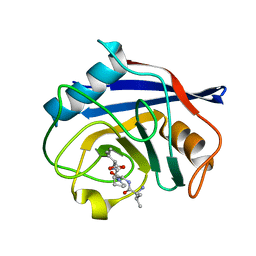 | | Structure of PPIA in complex with the peptide of NRF2 | | Descriptor: | NRF2 peptide, Peptidyl-prolyl cis-trans isomerase A, N-terminally processed | | Authors: | Wanyan, W, Hui, M, Jin, H, Lu, W. | | Deposit date: | 2023-01-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | PPIA dictates NRF2 stability to promote lung cancer progression.
Nat Commun, 15, 2024
|
|
6CUD
 
 | | Structure of the human TRPC3 in a lipid-occupied, closed state | | Descriptor: | (2R)-3-hydroxypropane-1,2-diyl dihexanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, W, Du, J, Fan, C, Choi, W. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human lipid-gated cation channel TRPC3.
Elife, 7, 2018
|
|
6WBK
 
 | |
6WBN
 
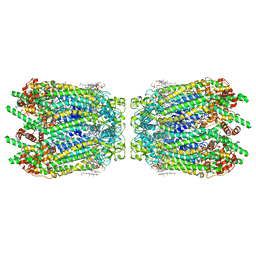 | | Cryo-EM structure of human Pannexin 1 channel N255A mutant, gap junction | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, DIACYL GLYCEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBM
 
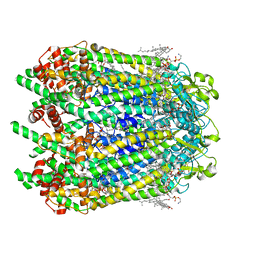 | | Cryo-EM structure of human Pannexin 1 channel N255A mutant | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, DIACYL GLYCEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBL
 
 | |
6WBG
 
 | | Cryo-EM structure of human Pannexin 1 channel with its C-terminal tail cleaved by caspase-7 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBF
 
 | | Cryo-EM structure of wild type human Pannexin 1 channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBI
 
 | |
7JNC
 
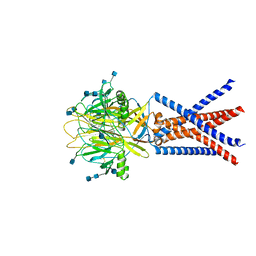 | | cryo-EM structure of human proton-activated chloride channel PAC at pH 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel | | Authors: | Lu, W, Ruan, R, Du, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-11-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures and pH-sensing mechanism of the proton-activated chloride channel.
Nature, 588, 2020
|
|
7JNA
 
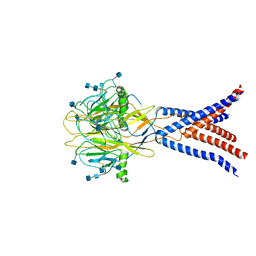 | | Cryo-EM structure of human proton-activated chloride channel PAC at pH 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel | | Authors: | Lu, W, Ruan, R, Du, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-11-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and pH-sensing mechanism of the proton-activated chloride channel.
Nature, 588, 2020
|
|
2KHT
 
 | | NMR Structure of human alpha defensin HNP-1 | | Descriptor: | Neutrophil defensin 1 | | Authors: | Zhang, Y, Li, S, Doherty, T.F, Lubkowski, J, Lu, W, Li, J, Barinka, C, Hong, M. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR.
J.Mol.Biol., 397, 2010
|
|
4E9A
 
 | | Structure of Peptide Deformylase form Helicobacter Pylori in complex with inhibitor | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COBALT (II) ION, ... | | Authors: | Cui, K, Zhu, L, Lu, W, Huang, J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Identification of Novel Peptide Deformylase Inhibitors from Natural Products
To be Published
|
|
1PN3
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-12 | | Release date: | 2003-08-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4YLW
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, D, Ouyang, P, Lu, W, Huang, J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound
To Be Published
|
|
1ZMQ
 
 | | Crystal structure of human alpha-defensin-6 | | Descriptor: | CHLORIDE ION, Defensin 6 | | Authors: | Lubkowski, J, Szyk, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human {alpha}-defensins HNP4, HD5, and HD6.
Protein Sci., 15, 2006
|
|
