3N34
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP, produced from 5-fluoro-6-azido-UMP | | Descriptor: | 1,2-ETHANEDIOL, 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2010-05-19 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP, produced from 5-fluoro-6-azido-UMP
To be Published
|
|
3MWA
 
 | |
3N3M
 
 | |
7RDQ
 
 | |
1RGQ
 
 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | Descriptor: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | Authors: | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
5T1G
 
 | | chromo shadow domain of CBX1 in complex with a histone peptide | | Descriptor: | Chromobox protein homolog 1, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | chromo shadow domain of CBX1 in complex with a histone peptide
To Be Published
|
|
5UK2
 
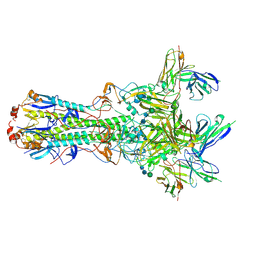 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UJZ
 
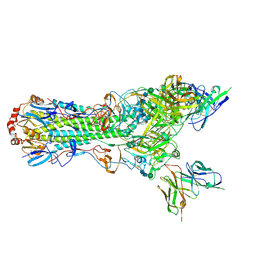 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UK0
 
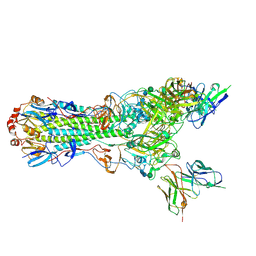 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
3DBP
 
 | | Crystal Structure of Human Orotidine 5'-Monophosphate Decarboxylase Complexed with 6-NH2-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Liu, Y, To, T, Bello, A.M, Kotra, L.P, Pai, E.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-Monophosphate Decarboxylase Complexed with 6-NH2-UMP
To be Published
|
|
5UZA
 
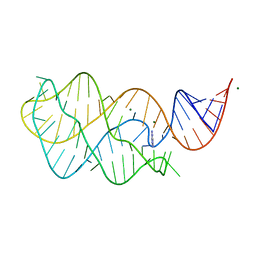 | | Adenine riboswitch aptamer domain labelled with iodo-uridine by position-selective labelling of RNA (PLOR) | | Descriptor: | ADENINE, MAGNESIUM ION, RNA (71-MER) | | Authors: | Liu, Y, Stagno, J.R, Wang, Y.-X. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA.
Nat Protoc, 13, 2018
|
|
5UK1
 
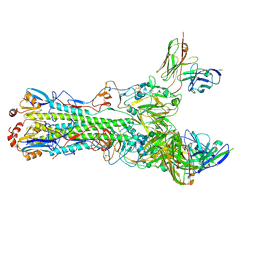 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
8DWH
 
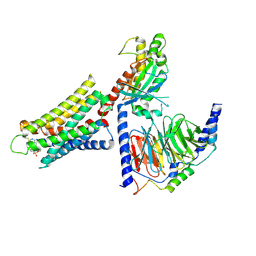 | | CryoEM structure of Gq-coupled MRGPRX1 with ligand Compound-16 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
8DWC
 
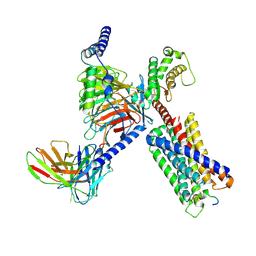 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
8DWG
 
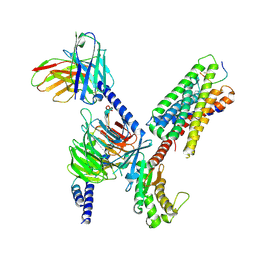 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382 | | Descriptor: | 2-[(cyclopropanesulfonyl)amino]-N-(2-ethoxyphenyl)benzamide, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
2AJI
 
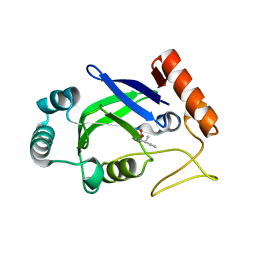 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with isoleucine | | Descriptor: | ISOLEUCINE, Leucyl-tRNA synthetase | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2AZP
 
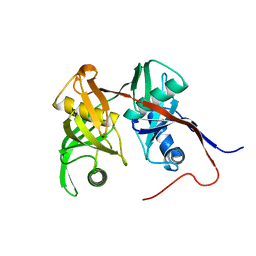 | | Crystal Structure of PA1268 Solved by Sulfur SAD | | Descriptor: | hypothetical protein PA1268 | | Authors: | Liu, Y, Gorodichtchenskaia, E, Skarina, T, Yang, C, Joachimiak, A, Edwards, A, Pai, E.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of PA1268 Solved by Sulfur SAD
To be Published
|
|
2AJG
 
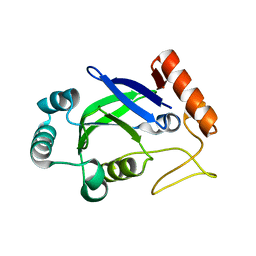 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase | | Descriptor: | Leucyl-tRNA synthetase | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-01 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
3SEC
 
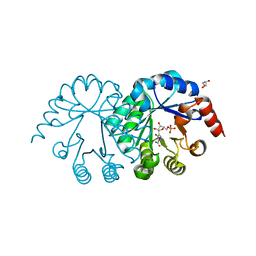 | |
2AJH
 
 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with methionine | | Descriptor: | Leucyl-tRNA synthetase, METHIONINE | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2EAW
 
 | |
2ESH
 
 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
4RR5
 
 | | The crystal structure of Synechocystis sp. PCC 6803 Malonyl-CoA: ACP Transacylase | | Descriptor: | ACETATE ION, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Liu, Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Structural and biochemical characterization of MCAT from photosynthetic microorganism Synechocystis sp. PCC 6803 reveal its stepwise catalytic mechanism
Biochem.Biophys.Res.Commun., 457, 2015
|
|
4M9E
 
 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-14 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
4F7H
 
 | | The crystal structure of kindlin-2 pleckstrin homology domain in free form | | Descriptor: | Fermitin family homolog 2, S,R MESO-TARTARIC ACID | | Authors: | Liu, Y, Zhu, Y, Qin, J, Ye, S, Zhang, R. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation.
Protein Cell, 3, 2012
|
|
