7YHL
 
 | |
7YGH
 
 | |
7YGL
 
 | |
8JBB
 
 | |
8JH1
 
 | |
8JBC
 
 | |
5XNP
 
 | | Crystal structures of human SALM5 in complex with human PTPdelta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.729 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5XNQ
 
 | | Crystal structures of human SALM5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5XME
 
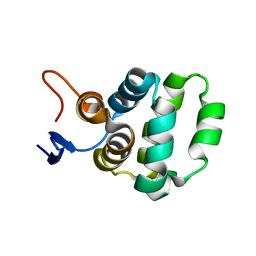 | | Solution structure of C-terminal domain of TRADD | | Descriptor: | Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2017-05-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Sci Rep, 7, 2017
|
|
5Z6Q
 
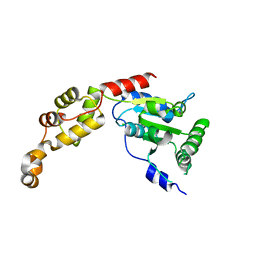 | | Crystal structure of AAA of Spastin | | Descriptor: | CHLORIDE ION, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The AAA protein spastin possesses two levels of basal ATPase activity
FEBS Lett., 592, 2018
|
|
5ZGG
 
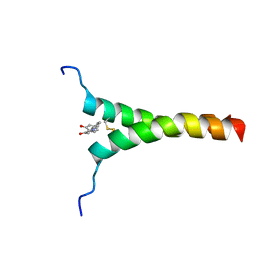 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
4XSK
 
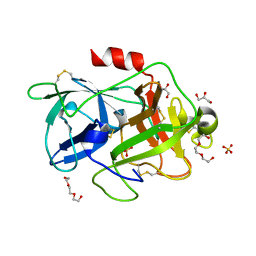 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
1GP7
 
 | |
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1CRW
 
 | |
2GIZ
 
 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
1JIA
 
 | |
1M8T
 
 | | Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Phospholipase a2 | | Authors: | Xu, S, Gu, L, Wang, Q, Shu, Y, Lin, Z. | | Deposit date: | 2002-07-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1BK9
 
 | | PHOSPHOLIPASE A2 MODIFIED BY PBPB | | Descriptor: | 1,4-BUTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Zhao, H, Tang, L, Wang, X, Lin, Z, Zhou, Y. | | Deposit date: | 1998-07-16 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a snake venom phospholipase A2 modified by p-bromo-phenacyl-bromide.
Toxicon, 36, 1998
|
|
4XKL
 
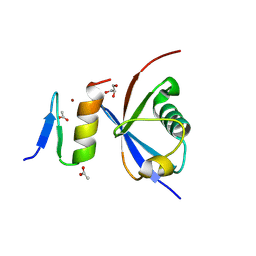 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
1HUO
 
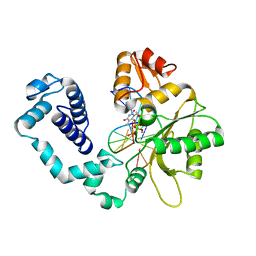 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1HUZ
 
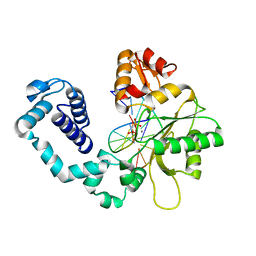 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1IJL
 
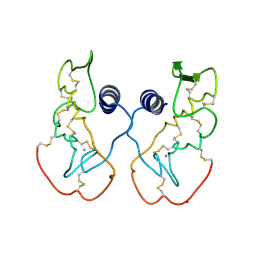 | | Crystal structure of acidic phospholipase A2 from deinagkistrodon acutus | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Gu, L, Zhang, H, Song, S, Zhou, Y, Lin, Z. | | Deposit date: | 2001-04-27 | | Release date: | 2001-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from the venom of Deinagkistrodon acutus.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2M0M
 
 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
