2N2Y
 
 | |
7Y77
 
 | | Crystal structure of rice NAL1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Yan, J.J, Guan, Z.Y, Yin, P, Xiong, L.Z. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Serine protease NAL1 exerts pleiotropic functions through degradation of TOPLESS-related corepressor in rice.
Nat.Plants, 9, 2023
|
|
6ABO
 
 | | human XRCC4 and IFFO1 complex | | Descriptor: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | Authors: | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | Deposit date: | 2018-07-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
8HXY
 
 | |
8HXX
 
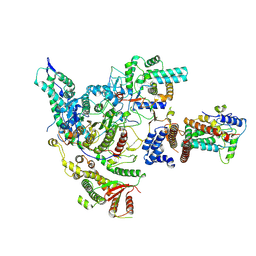 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HY0
 
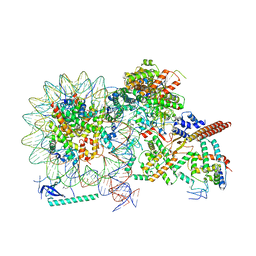 | |
8HXZ
 
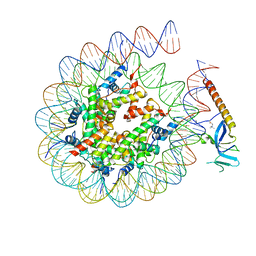 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8JHO
 
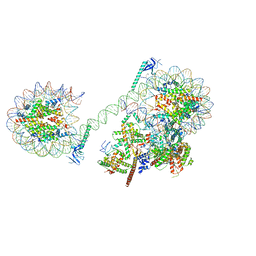 | |
6KMS
 
 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group I422) | | Descriptor: | Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYLMETHIONINE | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
6KMR
 
 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group P6122) | | Descriptor: | 1,2-ETHANEDIOL, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, ... | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
6JYM
 
 | |
8GK7
 
 | | MsbA bound to cerastecin C | | Descriptor: | 2-[(4-butylbenzene-1-sulfonyl)amino]-5-[(3-{4-[(4-butylbenzene-1-sulfonyl)amino]-3-carboxyanilino}-3-oxopropyl)carbamoyl]benzoic acid, Lipid A export ATP-binding/permease protein MsbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Y, Klein, D. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii
Nat Microbiol, 2024
|
|
6QFY
 
 | | CRYSTAL STRUCTURE OF PORCINE HEMAGGLUTINATING ENCEPHALOMYELITIS VIRUS SPIKE PROTEIN LECTIN DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Huizinga, E.G, Bakkers, M, Lang, Y. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
7R40
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 87G7 heavy chain variable region, ... | | Authors: | Hurdiss, D.L. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-20 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern.
Sci Immunol, 7, 2022
|
|
6WJH
 
 | |
6XES
 
 | | Tubulin-RB3_SLD in complex with compound 40a | | Descriptor: | DIMETHYL SULFOXIDE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6XER
 
 | | Tubulin-RB3_SLD in complex with colchicine | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6XET
 
 | | Tubulin-RB3_SLD in complex with compound 60c | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6YQQ
 
 | | ForT-PRPP complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ForT-PRPP complex, ... | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2020-04-18 | | Release date: | 2020-05-20 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex.
Chem.Commun.(Camb.), 56, 2020
|
|
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
