7XBO
 
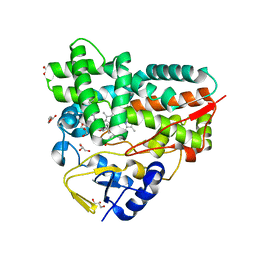 | | Crystal Structure of 10-dml-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBN
 
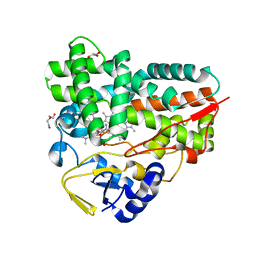 | | Crystal Structure of YC-17-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBM
 
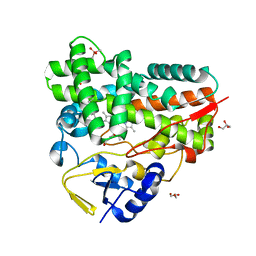 | | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | CACODYLATE ION, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238
Nat Commun, 2023
|
|
8ZAF
 
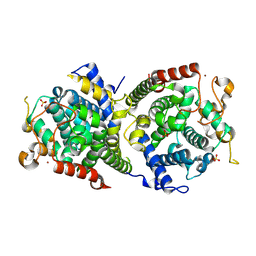 | | Crystal structure of SkABA3 from Shimazuella kribbensis | | Descriptor: | SULFATE ION, Sesquiterpene synthases, ZINC ION | | Authors: | Li, S.Y, Li, H, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular insights into a distinct class of terpenoid cyclases.
Nat Commun, 16, 2025
|
|
8ZAC
 
 | | Crystal structure of BcABA3 from Botrytis cinerea | | Descriptor: | Alpha-ionylideneethane synthase aba3, ZINC ION | | Authors: | Li, S.Y, Li, H, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular insights into a distinct class of terpenoid cyclases.
Nat Commun, 16, 2025
|
|
8ZAE
 
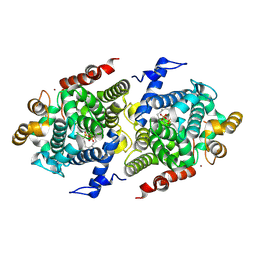 | | Crystal structure of RuABA3 from Rutstroemia sp. NJR-2017a WRK4 in complex with FsPP | | Descriptor: | Aba 3 protein, MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Li, S.Y, Li, H, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular insights into a distinct class of terpenoid cyclases.
Nat Commun, 16, 2025
|
|
8ZAG
 
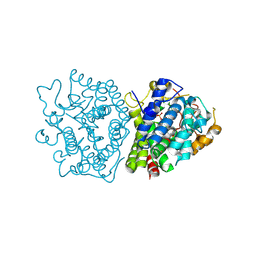 | | Crystal structure of SkABA3 from Shimazuella kribbensis in complex with PPi | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Sesquiterpene synthases, ... | | Authors: | Li, S.Y, Li, H, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into a distinct class of terpenoid cyclases.
Nat Commun, 16, 2025
|
|
8ZAD
 
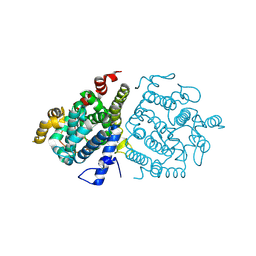 | | Crystal structure of RuABA3 from Rutstroemia sp. NJR-2017a WRK4 | | Descriptor: | Aba 3 protein, ZINC ION | | Authors: | Li, S.Y, Li, H, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular insights into a distinct class of terpenoid cyclases.
Nat Commun, 16, 2025
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
8W53
 
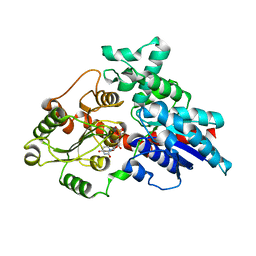 | | Crystal structure of LbUGT in complex with UDP | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Hu, D, Wang, G.Q. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.42816162 Å) | | Cite: | Functional and structural dissection of glycosyltransferases underlying the glycodiversity of wolfberry-derived bioactive ingredients lycibarbarspermidines.
Nat Commun, 15, 2024
|
|
8WP5
 
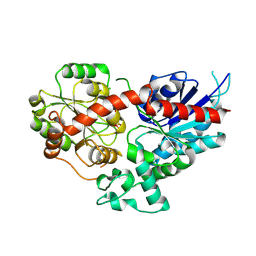 | | Crystal structure of LbUGT1 in complex with UDP | | Descriptor: | CHLORIDE ION, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Hu, D, Wang, G.Q. | | Deposit date: | 2023-10-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.57005954 Å) | | Cite: | Functional and structural dissection of glycosyltransferases underlying the glycodiversity of wolfberry-derived bioactive ingredients lycibarbarspermidines.
Nat Commun, 15, 2024
|
|
7DBI
 
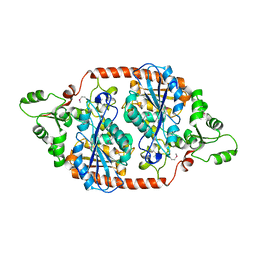 | | Crystal structure of the peroxisomal acyl-CoA hydrolase MpaH | | Descriptor: | acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
7DBL
 
 | | Acyl-CoA hydrolase MpaH' mutant S139A in complex with MPA | | Descriptor: | MYCOPHENOLIC ACID, acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
7QI8
 
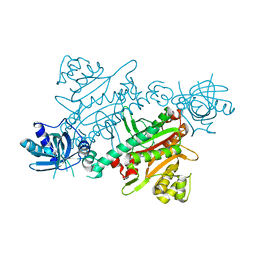 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE AND INHIBITOR | | Descriptor: | 2-azanyl-6-[(1~{S},7~{S})-2,2-bis(fluoranyl)-7-oxidanyl-cycloheptyl]-4-methoxy-7~{H}-pyrrolo[3,4-d]pyrimidin-5-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QHN
 
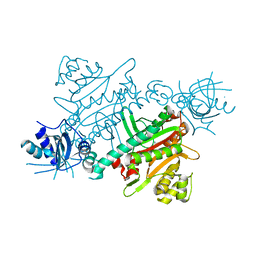 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE and an inhibitor | | Descriptor: | 6-azanyl-2-cyclohexyl-4-fluoranyl-1~{H}-pyrrolo[3,4-c]pyridin-3-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QH8
 
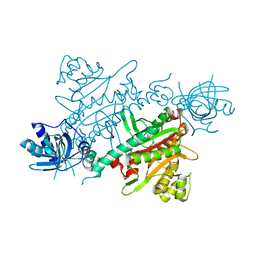 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
5YSW
 
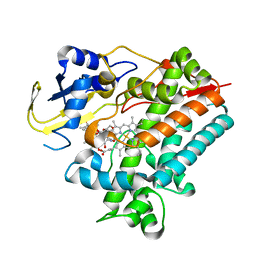 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
1T6W
 
 | | RATIONAL DESIGN OF A CALCIUM-BINDING ADHESION PROTEIN NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, hypothetical protein XP_346638 | | Authors: | Yang, W, Wilkins, A.L, Ye, Y, Liu, Z.-R, Urbauer, J.L, Kearney, A, van der Merwe, P.A, Yang, J.J. | | Deposit date: | 2004-05-07 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design of a calcium-binding protein with desired structure in a cell adhesion molecule.
J.Am.Chem.Soc., 127, 2005
|
|
3WJA
 
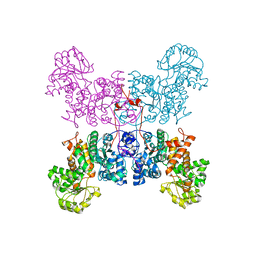 | | The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Li, S.-Y, Chen, M.-C, Yang, P.-C, Chan, N.-L, Liu, J.-H, Hung, H.-C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Structural characteristics of the nonallosteric human cytosolic malic enzyme.
Biochim.Biophys.Acta, 1844, 2014
|
|
6M4S
 
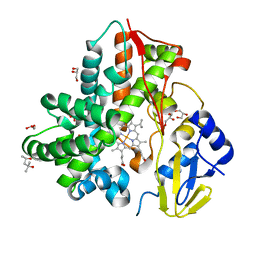 | | Crystal Structure Analysis of the cytochrome P450 CYP-Sb21 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cytochrome P450 hydroxylase sb21, ... | | Authors: | Li, F.W, Li, S.Y. | | Deposit date: | 2020-03-09 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided manipulation of the regioselectivity of the cyclosporine A hydroxylase CYP-sb21 from Sebekia benihana .
Synth Syst Biotechnol, 5, 2020
|
|
8YND
 
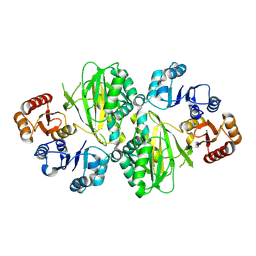 | | ATP-grasp peptide ligase from Streptomyces | | Descriptor: | ATP-grasp peptide ligase | | Authors: | Xu, M.J, Yan, Y.Q. | | Deposit date: | 2024-03-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of an ATP-Grasp Peptide-Ligase Producing Diverse Dipeptides Containing Unnatural Amino Acids
Acs Catalysis, 15, 2025
|
|
8YNQ
 
 | | ATP-grasp peptide ligase from Streptomyces | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Xu, M.J, Yan, Y.Q. | | Deposit date: | 2024-03-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of an ATP-Grasp Peptide-Ligase Producing Diverse Dipeptides Containing Unnatural Amino Acids
Acs Catalysis, 15, 2025
|
|
8GUE
 
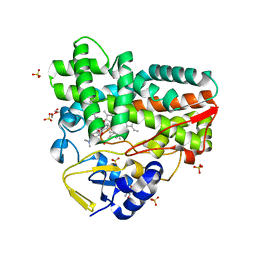 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-09-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
7CCF
 
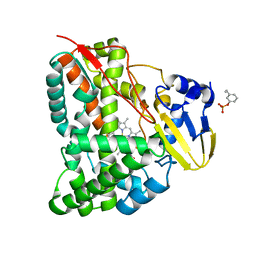 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | Descriptor: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
7CFS
 
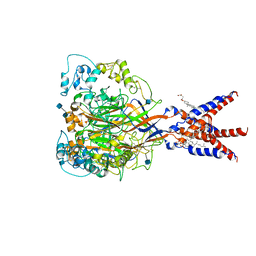 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
