7FIE
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7WSF
 
 | |
7WSE
 
 | |
7WSG
 
 | |
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
1YY8
 
 | | Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 | | Descriptor: | Cetuximab Fab Heavy chain, Cetuximab Fab Light chain | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
1YY9
 
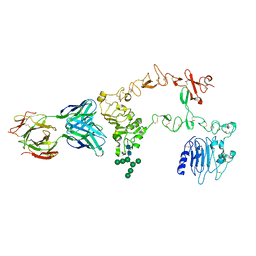 | | Structure of the extracellular domain of the epidermal growth factor receptor in complex with the Fab fragment of cetuximab/Erbitux/IMC-C225 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab Heavy chain, ... | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
2B4S
 
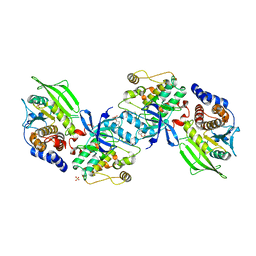 | | Crystal structure of a complex between PTP1B and the insulin receptor tyrosine kinase | | Descriptor: | Insulin receptor, SULFATE ION, Tyrosine-protein phosphatase, ... | | Authors: | Li, S, Depetris, R.S, Barford, D, Chernoff, J, Hubbard, S.R. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Complex between Protein Tyrosine Phosphatase 1B and the Insulin Receptor Tyrosine Kinase.
Structure, 13, 2005
|
|
5FXU
 
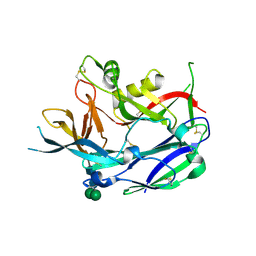 | | Crystal Structure of Puumala virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE POLYPROTEIN, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, S, Rissanen, I, Zeltina, A, Hepojoki, J, Raghwani, J, Harlos, K, Pybus, O.G, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Molecular-Level Account of the Antigenic Hantaviral Surface.
Cell Rep., 15, 2016
|
|
5FYN
 
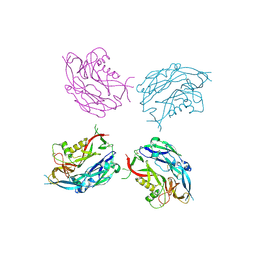 | | Sub-tomogram averaging of Tula virus glycoprotein spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PUUMALA VIRUS GN GLYCOPROTEIN, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, S, Rissanen, I, Zeltina, A, Hepojoki, J, Raghwani, J, Harlos, K, Pybus, O.G, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | A Molecular-Level Account of the Antigenic Hantaviral Surface.
Cell Rep., 15, 2016
|
|
2DOQ
 
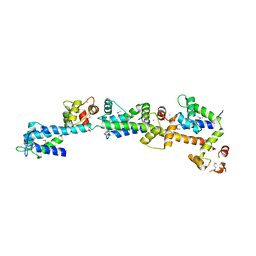 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, SFI1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
8IVI
 
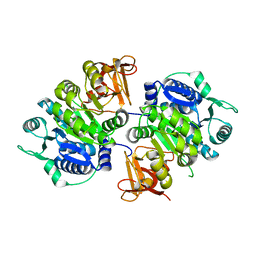 | | crystal structure of a medium-long chain fatty acyl-CoA ligase | | Descriptor: | Medium/long-chain-fatty-acid--CoA ligase FadD8 | | Authors: | Li, S. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural study of medium-long chain fatty acyl-CoA ligase FadD8 from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
3ZSN
 
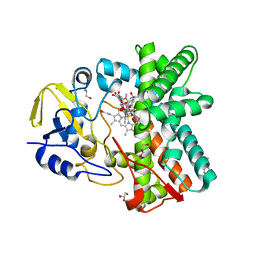 | | Structure of the mixed-function P450 MycG F286A mutant in complex with mycinamicin IV | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Anzai, Y, Kato, F, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-06-29 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
5YKR
 
 | |
5YKT
 
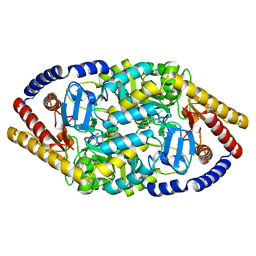 | |
8K3Y
 
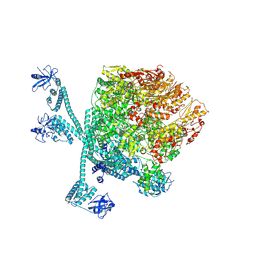 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
4H0N
 
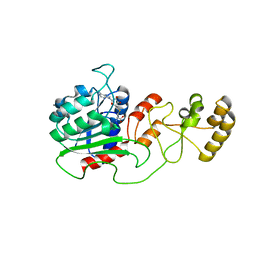 | | Crystal structure of Spodoptera frugiperda DNMT2 E260A/E261A/K263A mutant | | Descriptor: | CALCIUM ION, DNMT2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Li, S, Du, J, Yang, H, Yin, J, Zhong, J, Ding, J. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Functional and structural characterization of DNMT2 from Spodoptera frugiperda.
J Mol Cell Biol, 5, 2013
|
|
4I16
 
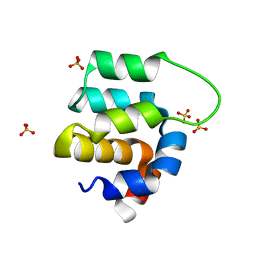 | | Crystal structure of CARMA1 CARD | | Descriptor: | Caspase recruitment domain-containing protein 11, SULFATE ION | | Authors: | Li, S, Yang, X, Shen, Y. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural insights into the assembly of CARMA1 and BCL10
Plos One, 7, 2012
|
|
6IKJ
 
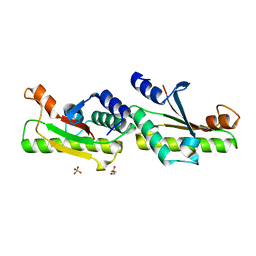 | | Crystal structure of YfiB(F48S) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IKK
 
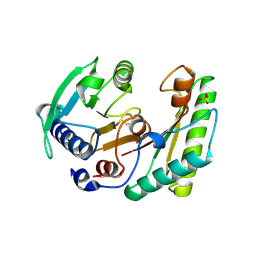 | |
6IKI
 
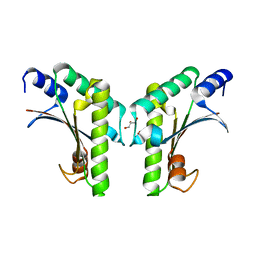 | | Crystal structure of YfiB(W55L) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
7YUU
 
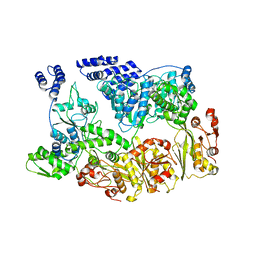 | | MtaLon-ADP for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUV
 
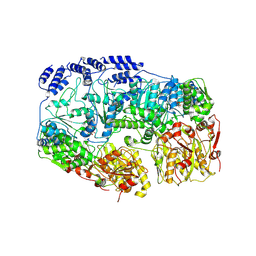 | | MtaLon-ADP for the spiral oligomers of tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUW
 
 | | MtaLon-ADP for the spiral oligomers of pentamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUX
 
 | | MtaLon-ADP for the spiral oligomers of hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
