6S4K
 
 | | The crystal structure of glycogen phosphorylase in complex with 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(4-phenyl-1,3-thiazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S4O
 
 | | The crystal structure of glycogen phosphorylase in complex with 9 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S4R
 
 | | The crystal structure of glycogen phosphorylase in complex with 11 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1,3-thiazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Papaioannou, O.S.E, Solovou, T.G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
4CTM
 
 | | Glucopyranosylidene-spiro-iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetic and Crystallographic Methods | | Descriptor: | (5R,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2-imino-6-oxa-1-thia-3-azaspiro[4.5]decan-4-one, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Alexacou, K.M, Papakonstantinou, M, Leonidas, D.D, Zographos, S.E, Chrysina, E.D. | | Deposit date: | 2014-03-15 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glucopyranosylidene-Spiro-Iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation for Inhibition of Glycogen Phosphorylase by Enzyme Kinetic and Crystallographic Methods.
Bioorg.Med.Chem., 22, 2014
|
|
4CTN
 
 | | Glucopyranosylidene-spiro-iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetic and Crystallographic Methods | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DIMETHYL SULFOXIDE, GLYCOGEN PHOSPHORYLASE, ... | | Authors: | Alexacou, K.M, Papakonstantinou, M, Leonidas, D.D, Zographos, S.E, Chrysina, E.D. | | Deposit date: | 2014-03-15 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glucopyranosylidene-Spiro-Iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation for Inhibition of Glycogen Phosphorylase by Enzyme Kinetic and Crystallographic Methods.
Bioorg.Med.Chem., 22, 2014
|
|
4CTO
 
 | | Glucopyranosylidene-spiro-iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetic and Crystallographic Methods | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(2Z,5R,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-4-oxo-6-oxa-1-thia-3-azaspiro[4.5]dec-2-ylidene]benzamide, ... | | Authors: | Alexacou, K.M, Papakonstantinou, M, Leonidas, D.D, Zographos, S.E, Chrysina, E.D. | | Deposit date: | 2014-03-15 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glucopyranosylidene-Spiro-Iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation for Inhibition of Glycogen Phosphorylase by Enzyme Kinetic and Crystallographic Methods.
Bioorg.Med.Chem., 22, 2014
|
|
4EL5
 
 | | Crystal structure of GPb in complex with DK12 | | Descriptor: | 5-ethynyl-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EJ2
 
 | | Crystal structure of GPb in complex with DK10 | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(hept-1-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EKE
 
 | | Crystal structure of GPb in complex with DK11 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-pentylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-09 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EKY
 
 | | Crystal structure of GPb in complex with DK15 | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(pent-1-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EL0
 
 | | Crystal structure of GPb in complex with DK16 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-propylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4G8Y
 
 | | Crystal structure of Ribonuclease A in complex with 5b | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4G90
 
 | | Crystal structure of Ribonuclease A in complex with 5e | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-fluoro-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4G9M
 
 | |
4G8V
 
 | | Crystal structure of Ribonuclease A in complex with 5a | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4G9N
 
 | |
1FZV
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PLACENTA GROWTH FACTOR-1 (PLGF-1), AN ANGIOGENIC PROTEIN AT 2.0A RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PLACENTA GROWTH FACTOR | | Authors: | Iyer, S, Leonidas, D.D, Swaminathan, G.J, Maglione, D, Battisti, M, Tucci, M, Persico, M.G, Acharya, K.R. | | Deposit date: | 2000-10-04 | | Release date: | 2001-05-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution.
J.Biol.Chem., 276, 2001
|
|
1GG8
 
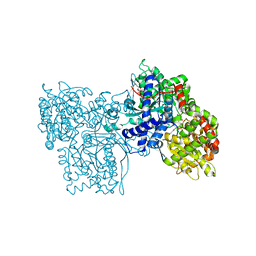 | | DESIGN OF INHIBITORS OF GLYCOGEN PHOSPHORYLASE: A STUDY OF ALPHA-AND BETA-C-GLUCOSIDES AND 1-THIO-BETA-D-GLUCOSE COMPOUNDS | | Descriptor: | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Watson, K.A, Mitchell, E.P, Johnson, L.N, Son, J.C, Bichard, C.J, Orchard, M.G, Fleet, G.W, Oikonomakos, N.G, Leonidas, D.D, Kontou, M, Papageorgiou, A.C. | | Deposit date: | 2000-07-30 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design of inhibitors of glycogen phosphorylase: a study of alpha- and beta-C-glucosides and 1-thio-beta-D-glucose compounds.
Biochemistry, 33, 1994
|
|
1H8U
 
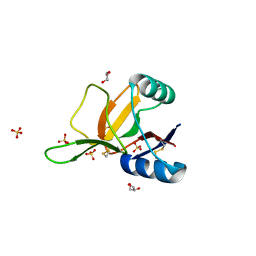 | | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A: An Atypical Lectin with a Paradigm Shift in Specificity | | Descriptor: | EOSINOPHIL GRANULE MAJOR BASIC PROTEIN 1, GLYCEROL, SULFATE ION | | Authors: | Swaminathan, G.J, Weaver, A.J, Loegering, D.A, Checkel, J.L, Leonidas, D.D, Gleich, G.J, Acharya, K.R. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A. An Atypical Lectin with a Paradigm Shift in Specificity
J.Biol.Chem., 276, 2001
|
|
1GV7
 
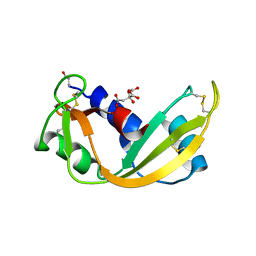 | | ARH-I, an angiogenin/RNase A chimera | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Shapiro, R, Hares, M.C, Leonidas, D.D, Acharya, K.R. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Guest-Host Crosstalk in an Angiogenin-RNase A Chimeric Protein
Biochemistry, 41, 2002
|
|
2FET
 
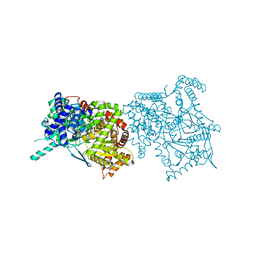 | | Synthesis of C-D-Glycopyranosyl-Hydroquinones and-Benzoquinones. Inhibition of PTP1B. Inhibition of and binding to glycogen phosphorylase in the crystal | | Descriptor: | (1S)-1,5-anhydro-1-(2,5-dihydroxyphenyl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2005-12-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | In the Search of Glycogen Phosphorylase Inhibitors: Synthesis of C-D-Glycopyranosylbenzo(hydro)quinones Inhibition of and Binding to Glycogen Phosphorylase in the Crystal
Eur.J.Org.Chem., 4, 2007
|
|
2F3Q
 
 | |
2F3P
 
 | |
2F3U
 
 | |
2F3S
 
 | |
