6V55
 
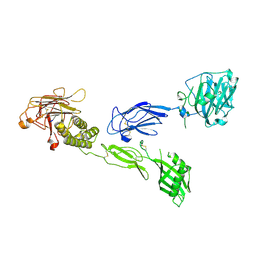 | | Full extracellular region of zebrafish Gpr126/Adgrg6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor G6, CALCIUM ION | | Authors: | Leon, K, Arac, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for adhesion G protein-coupled receptor Gpr126 function.
Nat Commun, 11, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4GZZ
 
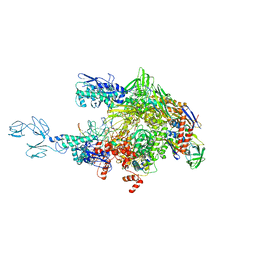 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.2927 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4GZY
 
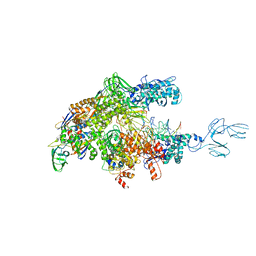 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5054 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
1M0U
 
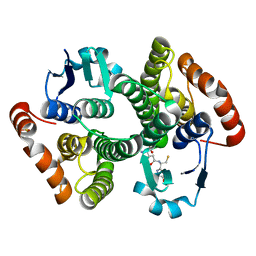 | | Crystal Structure of the Drosophila Glutathione S-transferase-2 in Complex with Glutathione | | Descriptor: | GLUTATHIONE, GST2 gene product, SULFATE ION | | Authors: | Agianian, B, Tucker, P.A, Schouten, A, Leonard, K, Bullard, B, Gros, P. | | Deposit date: | 2002-06-14 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Drosophila Sigma Class Glutathione S-transferase Reveals a Novel
Active Site Topography Suited for Lipid Peroxidation Products
J.Mol.Biol., 326, 2003
|
|
5VT0
 
 | |
8EM3
 
 | |
6VHH
 
 | | Human Teneurin-2 and human Latrophilin-3 binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L3, ... | | Authors: | Xie, Y, Li, J, Arac, D, Zhao, M. | | Deposit date: | 2020-01-09 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism.
Nat Commun, 11, 2020
|
|
8DJG
 
 | | ADGRL3-lectin domain in complex with an activating synthetic antibody fragment | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isoform 2 of Adhesion G protein-coupled receptor L3, ... | | Authors: | Kordon, S.P, Bandekar, S.J, Arac, D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Isoform- and ligand-specific modulation of the adhesion GPCR ADGRL3/Latrophilin3 by a synthetic binder.
Nat Commun, 14, 2023
|
|
4XLS
 
 | |
4XLR
 
 | |
4XAX
 
 | | Crystal structure of Thermus thermophilus CarD in complex with the Thermus aquaticus RNA polymerase beta1 domain | | Descriptor: | 1,2-ETHANEDIOL, CarD, DNA-directed RNA polymerase subunit beta domain 1 | | Authors: | Chen, J, Bae, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex.
Elife, 4, 2015
|
|
5KVM
 
 | | Extracellular region of mouse GPR56/ADGRG1 in complex with FN3 monobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor G1, ... | | Authors: | Salzman, G.S, Ding, C, Koide, S, Arac, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structural Basis for Regulation of GPR56/ADGRG1 by Its Alternatively Spliced Extracellular Domains.
Neuron, 91, 2016
|
|
6CMX
 
 | | Human Teneurin 2 extra-cellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | Authors: | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
4L5G
 
 | | Crystal structure of Thermus thermophilus CarD | | Descriptor: | CarD | | Authors: | Srivastava, D.B, Westblade, L.F, Campbell, E.A, Darst, S.A. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.3902 Å) | | Cite: | Structure and function of CarD, an essential mycobacterial transcription factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6VSW
 
 | | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of RORgt | | Descriptor: | 5-(2,3-dichloro-4-{[(2S)-1,1,1-trifluoropropan-2-yl]sulfamoyl}phenyl)-4-(4-fluoropiperidine-1-carbonyl)-N-(2-hydroxy-2-methylpropyl)-1,3-thiazole-2-carboxamide, RAR-related orphan receptor C | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of ROR gamma t.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6NAD
 
 | |
5UE4
 
 | | proMMP-9desFnII complexed to JNJ0966 INHIBITOR | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, SULFATE ION, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
5UE3
 
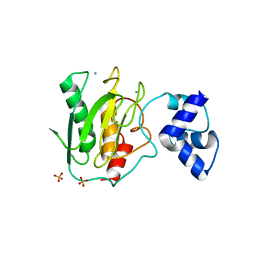 | | proMMP-9desFnII | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Matrix metalloproteinase-9, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
5IGM
 
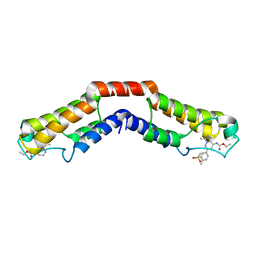 | | Crystal structure of the bromodomain of human BRD9 in complex with bromosporine (BSP) | | Descriptor: | Bromodomain-containing protein 9, Bromosporine | | Authors: | Tallant, C, Filippakopoulos, P, Picaud, S, Nunez-Alonso, G, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
5IGL
 
 | | Crystal structure of the second bromodomain of human TAF1L in complex with bromosporine (BSP) | | Descriptor: | Bromosporine, Transcription initiation factor TFIID subunit 1-like | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Krojer, T, Tallant, C, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
5IGK
 
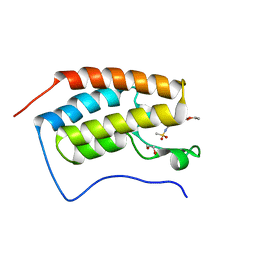 | | Crystal structure of the first bromodomain of human BRD4 in complex with bromosporine (BSP) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
5UFO
 
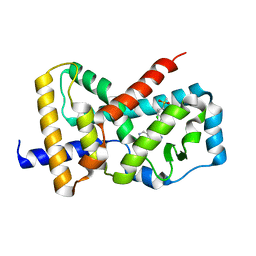 | | Structure of RORgt bound to | | Descriptor: | (S)-{4-chloro-2-methoxy-3-[4-(methylsulfonyl)phenyl]quinolin-6-yl}(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
