5NHC
 
 | |
5NH4
 
 | |
5NHD
 
 | |
5OHU
 
 | |
8VB1
 
 | | Crystal structure of HIV-1 protease with GS-9770 | | 分子名称: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | 著者 | Lansdon, E.B. | | 登録日 | 2023-12-11 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
4UW2
 
 | | Crystal structure of Csm1 in T.onnurineus | | 分子名称: | CSM1 | | 著者 | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | 登録日 | 2014-08-08 | | 公開日 | 2015-03-25 | | 最終更新日 | 2015-09-23 | | 実験手法 | X-RAY DIFFRACTION (2.632 Å) | | 主引用文献 | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
1E9Z
 
 | |
1E9Y
 
 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | 分子名称: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | 著者 | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | 登録日 | 2000-11-01 | | 公開日 | 2001-11-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | 分子名称: | Conserved hypothetical secreted protein, GLYCEROL | | 著者 | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | 登録日 | 2015-02-05 | | 公開日 | 2015-09-02 | | 最終更新日 | 2022-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
4Y4V
 
 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | 分子名称: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | 著者 | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | 登録日 | 2015-02-11 | | 公開日 | 2015-09-02 | | 最終更新日 | 2022-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
7QAJ
 
 | | ZK002 with Anti-angiogenic and Anti-inflamamtory Properties | | 分子名称: | SULFATE ION, Snaclec clone 2100755 alpha, Snaclec clone 2100755 beta | | 著者 | Wong, W.Y, Chan, B.D, Muk Lan Lee, M, Dai, X, Tsim, K.W.K, Hsiao, W.L.W, Li, M, Li, X.Y, Tai, W.C.S. | | 登録日 | 2021-11-17 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Isolation and characterization of ZK002, a novel dual function snake venom protein from Deinagkistrodon acutus with anti-angiogenic and anti-inflammatory properties.
Front Pharmacol, 14, 2023
|
|
7O4C
 
 | |
7OK9
 
 | |
7O4A
 
 | |
7O49
 
 | |
7O4B
 
 | |
8C54
 
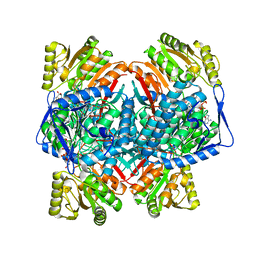 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | 著者 | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | 登録日 | 2023-01-06 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
6TW3
 
 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | 分子名称: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | 著者 | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.352 Å) | | 主引用文献 | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW9
 
 | | HumRadA22F in complex with CAM833 | | 分子名称: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | 著者 | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW4
 
 | | HumRadA22F in complex with compound 6 | | 分子名称: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | 著者 | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TV3
 
 | | HumRadA1 in complex with 3-amino-2-naphthoic acid | | 分子名称: | 3-azanylnaphthalene-2-carboxylic acid, DNA repair and recombination protein RadA, GLYCEROL, ... | | 著者 | Marsh, M.E, Hyvonen, M. | | 登録日 | 2020-01-08 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | 分子名称: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | 著者 | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | 登録日 | 2020-01-16 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
3TD5
 
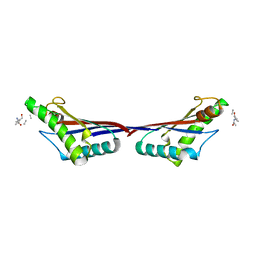 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | 分子名称: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | 著者 | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | 登録日 | 2011-08-10 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
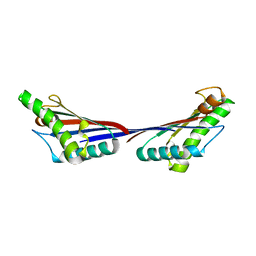 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | 分子名称: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | 著者 | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | 登録日 | 2011-08-10 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
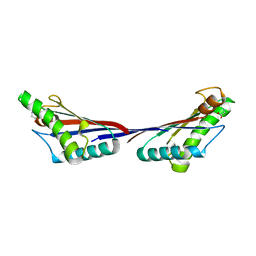 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | 分子名称: | GLYCINE, Outer membrane protein omp38 | | 著者 | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | 登録日 | 2011-08-10 | | 公開日 | 2011-10-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
