1LVG
 
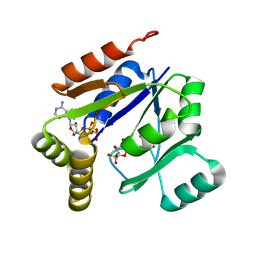 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
2HAZ
 
 | | Crystal structure of the first fibronectin domain of human NCAM1 | | Descriptor: | Neural cell adhesion molecule 1, SODIUM ION | | Authors: | Sekulic, N, Lavie, A. | | Deposit date: | 2006-06-13 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel alpha-helix in the first fibronectin type III repeat of the neural cell adhesion molecule is critical for N-glycan polysialylation.
J.Biol.Chem., 281, 2006
|
|
5DND
 
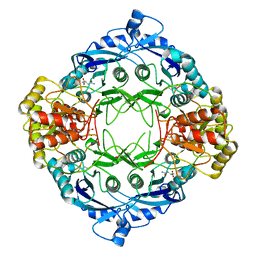 | |
5DNE
 
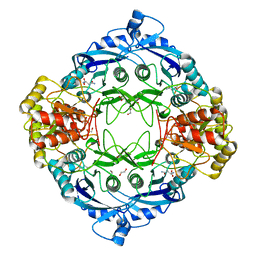 | |
5DNC
 
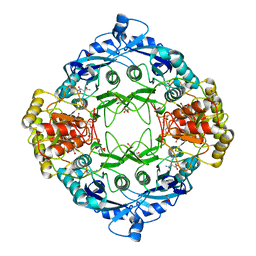 | |
1XYM
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | D-glucose, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
1XYL
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
6AQB
 
 | |
6C4S
 
 | |
6C4U
 
 | | Engineered FHA with Myc-pTBD peptide | | Descriptor: | Forkhead-associated 1, GLYCEROL, Myc-pTBD peptide | | Authors: | Kall, S.L, Lavie, A. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Generating a recombinant phosphothreonine-binding domain for a phosphopeptide of the human transcription factor, c-Myc.
N Biotechnol, 45, 2018
|
|
6CQ7
 
 | |
5K28
 
 | | Structure of the unbound SH3 domain of MLK3 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 11 | | Authors: | Kall, S.K, Lavie, A. | | Deposit date: | 2016-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of two distinct peptide-binding pockets in the SH3 domain of human mixed-lineage kinase 3.
J. Biol. Chem., 293, 2018
|
|
5K26
 
 | |
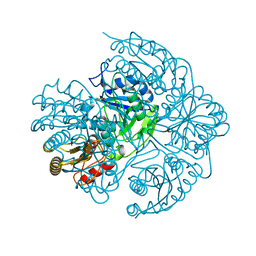
5HW0
 
 | |
5I4B
 
 | |
3HP1
 
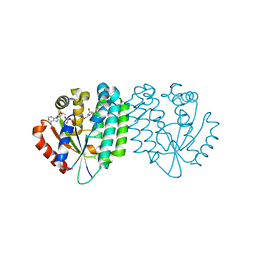 | |
5I3Z
 
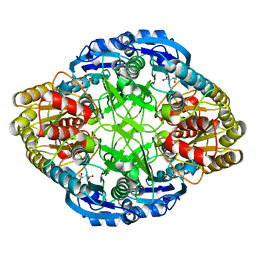 | |
5I48
 
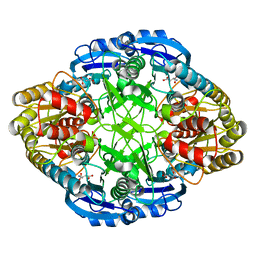 | |
5F52
 
 | | Erwinia chrysanthemi L-asparaginase + Aspartic acid | | Descriptor: | ASPARTIC ACID, DI(HYDROXYETHYL)ETHER, L-asparaginase | | Authors: | Nguyen, H.A, Lavie, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Insight into Substrate Selectivity of Erwinia chrysanthemi l-Asparaginase.
Biochemistry, 55, 2016
|
|
3KFX
 
 | | Human dCK complex with 5-Me dC and ADP | | Descriptor: | 5-METHYL-2'-DEOXYCYTIDINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase | | Authors: | Hazra, S, Lavie, A. | | Deposit date: | 2009-10-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and kinetic characterization of human deoxycytidine kinase variants able to phosphorylate 5-substituted deoxycytidine and thymidine analogues .
Biochemistry, 49, 2010
|
|
2A2Z
 
 | | Crystal Structure of human deoxycytidine kinase in complex with deoxycytidine and uridine diphosphate | | Descriptor: | 2'-DEOXYCYTIDINE, CALCIUM ION, Deoxycytidine kinase, ... | | Authors: | Godsey, M.H, Ort, S, Sabini, E, Konrad, M, Lavie, A. | | Deposit date: | 2005-06-23 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the preference of UTP over ATP in human deoxycytidine kinase: illuminating the role of main-chain reorganization.
Biochemistry, 45, 2006
|
|
4PVQ
 
 | | Crystal structure of sulfate-bound human l-asparaginase protein | | Descriptor: | IODIDE ION, Isoaspartyl peptidase/L-asparaginase, SODIUM ION, ... | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structures of apo and product-bound human L-asparaginase: insights into the mechanism of autoproteolysis and substrate hydrolysis.
Biochemistry, 51, 2012
|
|
4Q1B
 
 | |
4Q1D
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 9 {2-{[(1R)-1-{2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-propyl-1,3-thiazol-4-yl}ethyl]sulfanyl}pyrimidine-4,6-diamine} | | Descriptor: | (R)-2-((1-(2-(3-(2-fluoroethoxy)-4-methoxyphenyl)-5-propylthiazol-4-yl)ethyl)thio)pyrimidine-4,6-diamine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
4PVP
 
 | | Crystal structure of malonate-bound human L-asparaginase protein | | Descriptor: | Isoaspartyl peptidase/L-asparaginase, MALONATE ION, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of apo and product-bound human L-asparaginase: insights into the mechanism of autoproteolysis and substrate hydrolysis.
Biochemistry, 51, 2012
|
|
