4V9C
 
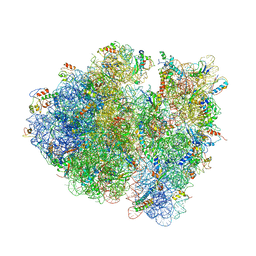 | | Allosteric control of the ribosome by small-molecule antibiotics | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Cate, J.H.D, Pulk, A, Blanchard, S.C, Wang, L, Feldman, M.B, Wasserman, M.R, Altman, R. | | Deposit date: | 2012-07-25 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Allosteric control of the ribosome by small-molecule antibiotics.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1G7Y
 
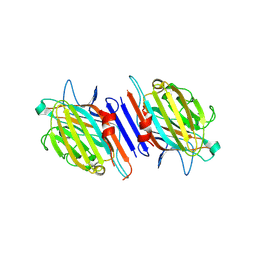 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | Authors: | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
1G7C
 
 | | YEAST EEF1A:EEF1BA IN COMPLEX WITH GDPNP | | Descriptor: | ELONGATION FACTOR 1-ALPHA, ELONGATION FACTOR 1-BETA, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
1H6O
 
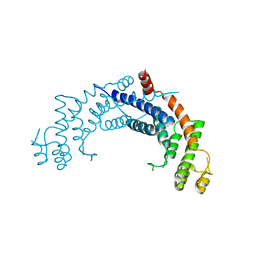 | | Dimerisation domain from human TRF1 | | Descriptor: | TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Fairall, L, Chapman, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2.
Mol.Cell, 8, 2001
|
|
1H6P
 
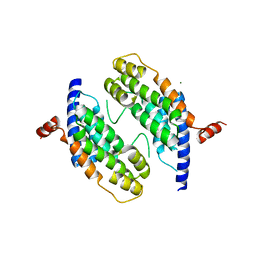 | | Dimeristion domain from human TRF2 | | Descriptor: | MAGNESIUM ION, TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Chapman, L, Fairall, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Trfh Dimerization Domain of the Human Telomere Proteins Trf1 and Trf2
Mol.Cell, 8, 2001
|
|
1HOY
 
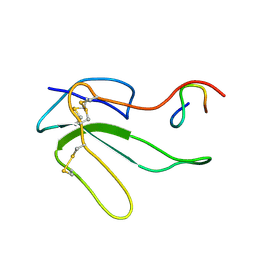 | | NMR STRUCTURE OF THE COMPLEX BETWEEN A-BUNGAROTOXIN AND A MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Niccolai, N, Neri, P. | | Deposit date: | 2000-12-12 | | Release date: | 2000-12-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1IJE
 
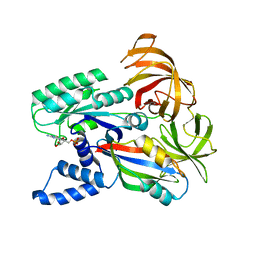 | | Nucleotide Exchange Intermediates in the eEF1A-eEF1Ba Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, elongation factor 1-alpha, elongation factor 1-beta | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
1IJF
 
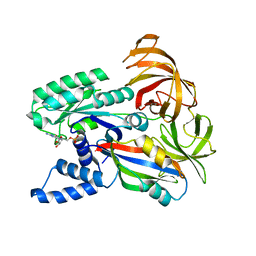 | | Nucleotide exchange mechanisms in the eEF1A-eEF1Ba complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, elongation factor 1-alpha, ... | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
3B6E
 
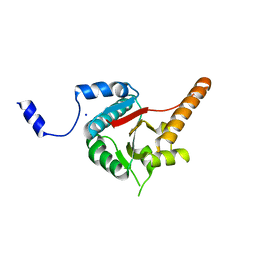 | | Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SODIUM ION | | Authors: | Karlberg, T, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain.
To be Published
|
|
4UUQ
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
5FCS
 
 | | Diabody | | Descriptor: | Diabody, SULFATE ION | | Authors: | Mosyak, L, Root, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-14 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of PF-06671008, a Highly Potent Anti-P-cadherin/Anti-CD3 Bispecific DART Molecule with Extended Half-Life for the Treatment of Cancer.
Antibodies, 5, 2016
|
|
1AMH
 
 | | UNCOMPLEXED RAT TRYPSIN MUTANT WITH ASP 189 REPLACED WITH SER (D189S) | | Descriptor: | ANIONIC TRYPSIN, CALCIUM ION | | Authors: | Szabo, E, Bocskei, Z.S, Naray-Szabo, G, Graf, L. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of Asp189Ser trypsin provides evidence for an inherent structural plasticity of the protease.
Eur.J.Biochem., 263, 1999
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
4X2Z
 
 | | Structural view and substrate specificity of papain-like protease from Avian Infectious Bronchitis Virus | | Descriptor: | Nonstructural protein 3, ZINC ION | | Authors: | Kong, L, Shaw, N, Yan, L, Lou, Z, Rao, Z. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-28 | | Last modified: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural View and Substrate Specificity of Papain-like Protease from Avian Infectious Bronchitis Virus.
J.Biol.Chem., 290, 2015
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
4XOU
 
 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
4Y0D
 
 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
7S5B
 
 | |
1AYO
 
 | | RECEPTOR BINDING DOMAIN OF BOVINE ALPHA-2-MACROGLOBULIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-2-MACROGLOBULIN, CALCIUM ION | | Authors: | Jenner, L.B, Husted, L, Thirup, S, Sottrup-Jensen, L, Nyborg, J. | | Deposit date: | 1997-11-07 | | Release date: | 1998-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the receptor-binding domain of alpha 2-macroglobulin.
Structure, 6, 1998
|
|
4Y0H
 
 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | 4-aminobutyrate aminotransferase, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-06 | | Release date: | 2015-03-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
