5KZY
 
 | |
5KZZ
 
 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in its apo-form | | Descriptor: | ACETATE ION, GLYCEROL, Smoothened, ... | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
1CEK
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE MEMBRANE-EMBEDDED M2 CHANNEL-LINING SEGMENT FROM THE NICOTINIC ACETYLCHOLINE RECEPTOR BY SOLID-STATE NMR SPECTROSCOPY | | Descriptor: | PROTEIN (ACETYLCHOLINE RECEPTOR M2) | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
6LHS
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHU
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHV
 
 | | Structure of FANCA and FANCG Complex | | Descriptor: | Fanconi anemia complementation group A, Fanconi anemia complementation group G | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6D35
 
 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | Descriptor: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6D32
 
 | | Crystal structure of Xenopus Smoothened in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6E3S
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist PT2385 | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3T
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist T1001 | | Descriptor: | (6S)-6-(4-bromophenyl)-2,3,5,6-tetrahydroimidazo[2,1-b][1,3]thiazole, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3U
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Agonist M1001 | | Descriptor: | 3-{[2-(pyrrolidin-1-yl)phenyl]amino}-1H-1lambda~6~,2-benzothiazole-1,1-dione, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
6W5K
 
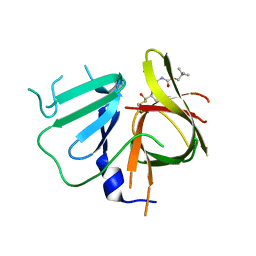 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | Descriptor: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
6WCF
 
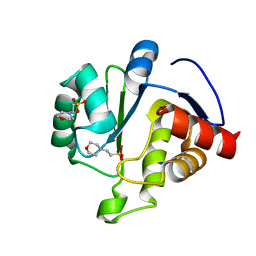 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.065 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WTC
 
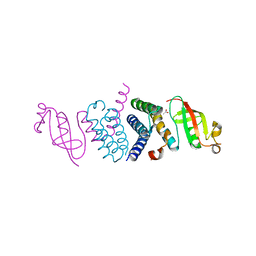 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
2XMQ
 
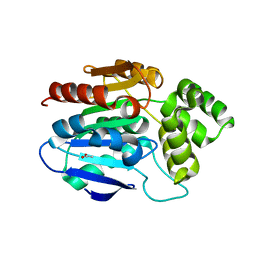 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
6WEN
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | Descriptor: | CHLORIDE ION, Non-structural protein 3 | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
2XMR
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
6WRH
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
2XMS
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
4X3Z
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
6XMK
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
4XBD
 
 | | 1.45A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Orthorhombic P Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
