6IJ4
 
 | |
6J2V
 
 | | GABA aminotransferase from Corynebacterium glutamicum | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GLYCEROL, PLP-dependent aminotransferases | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase in complex with a PLP-GABA adduct from Corynebacterium glutamicum.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6LRP
 
 | |
6LRT
 
 | |
8I4N
 
 | |
8I4Q
 
 | |
5M47
 
 | |
8HJW
 
 | | Bi-functional malonyl-CoA reductuase from Chloroflexus aurantiacus | | Descriptor: | Short-chain dehydrogenase/reductase SDR | | Authors: | Ahn, J.W, Kim, S. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of bifunctional malonyl-CoA reductase from Chloroflexus aurantiacus reveals a dynamic domain movement for high enzymatic activity.
Int.J.Biol.Macromol., 242, 2023
|
|
2XMQ
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMS
 
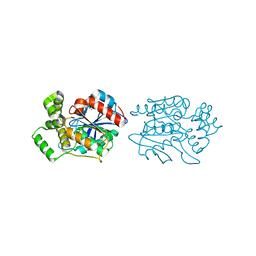 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMR
 
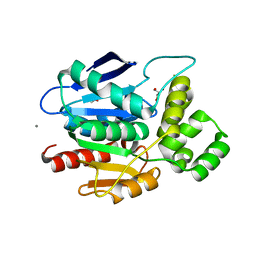 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
3EGM
 
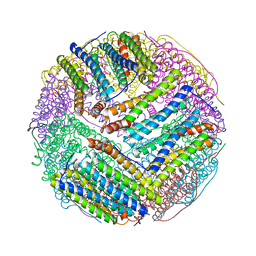 | | Structural basis of iron transport gating in Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Shin, H.J, Lee, J.H. | | Deposit date: | 2008-09-11 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of ferritin from Helicobacter pylori reveals unusual conformational changes for iron uptake.
J.Mol.Biol., 390, 2009
|
|
4REP
 
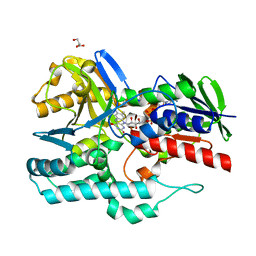 | | Crystal Structure of gamma-carotenoid desaturase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Gamma-carotene desaturase | | Authors: | Ahn, J.-W, Kim, E.-J, Kim, S, Kim, K.-J. | | Deposit date: | 2014-09-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of 1'-OH-carotenoid 3,4-desaturase from Nonlabens dokdonensis DSW-6.
Enzyme.Microb.Technol., 77, 2015
|
|
2XVJ
 
 | |
2XVH
 
 | |
2XVF
 
 | | Crystal structure of bacterial flavin-containing monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVIN-CONTAINING MONOOXYGENASE, GLYCEROL, ... | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2010-10-26 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analysis of Bacterial Flavin-Containing Monooxygenase Reveals its Ping-Pong-Type Reaction Mechanism.
J.Struct.Biol., 175, 2011
|
|
2XVE
 
 | | Crystal structure of bacterial flavin-containing monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVIN-CONTAINING MONOOXYGENASE, GLYCEROL, ... | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2010-10-26 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Analysis of Bacterial Flavin-Containing Monooxygenase Reveals its Ping-Pong-Type Reaction Mechanism.
J.Struct.Biol., 175, 2011
|
|
2XVI
 
 | |
7VG5
 
 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
7VG4
 
 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 strain | | Descriptor: | Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
7BYP
 
 | | Lysozyme structure SASE1 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7BYO
 
 | | Lysozyme structure SS1 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D01
 
 | | Lysozyme structure SS2 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D04
 
 | | Lysozyme structure SS3 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D05
 
 | | Lysozyme structure SASE3 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
