3AKK
 
 | |
3AKL
 
 | |
4XGS
 
 | |
2QKY
 
 | | complex structure of dipeptidyl peptidase IV and a oxadiazolyl ketone | | Descriptor: | 2-[(2-{(2S,4S)-2-[(R)-(5-tert-butyl-1,3,4-oxadiazol-2-yl)(hydroxy)methyl]-4-fluoropyrrolidin-1-yl}-2-oxoethyl)amino]-2-methylpropan-1-ol, Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) (Dipeptidyl peptidase 4 soluble form) (Dipeptidyl peptidase IV soluble form) | | Authors: | Kim, K.-H, Hong, S.Y, Koo, K.D, Lee, C.-S, Kim, G.T, Han, H.O. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, SAR, and X-ray structure of novel potent DPPIV inhibitors: oxadiazolyl ketones.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4YJI
 
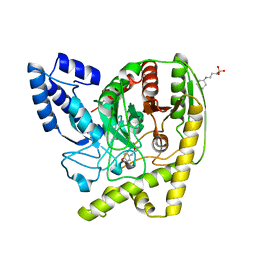 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Aryl acylamidase, N-(4-HYDROXYPHENYL)ACETAMIDE (TYLENOL) | | Authors: | Choi, I.-G, Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
4YJ6
 
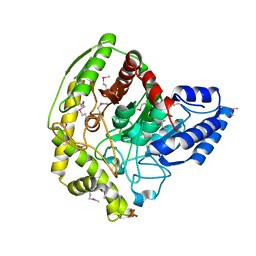 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | Aryl acylamidase, PHOSPHATE ION | | Authors: | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
3G6N
 
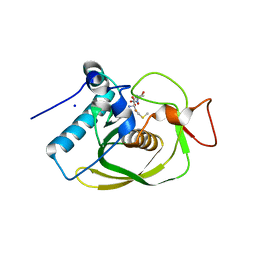 | | Crystal structure of an EfPDF complex with Met-Ala-Ser | | Descriptor: | FE (III) ION, Peptide deformylase, SODIUM ION, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
8GTI
 
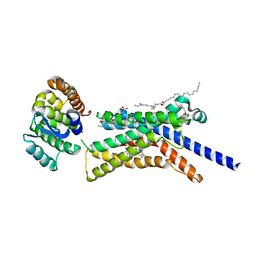 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTG
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
2OWY
 
 | |
2GJN
 
 | |
2GJL
 
 | | Crystal Structure of 2-nitropropane dioxygenase | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein PA1024 | | Authors: | Suh, S.W. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 2-Nitropropane Dioxygenase Complexed with FMN and Substrate: identification of the catalytic base
J.Biol.Chem., 281, 2006
|
|
3W8L
 
 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6CDD
 
 | |
7DB5
 
 | | Crystal structure of alpha-L-fucosidase from Vibrio sp. strain EJY3 | | Descriptor: | Alpha-L-fucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Hong, H, Kim, K.-J. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dual alpha-1,4- and beta-1,4-Glycosidase Activities by the Novel Carbohydrate-Binding Module in alpha-l-Fucosidase from Vibrio sp. Strain EJY3.
J.Agric.Food Chem., 69, 2021
|
|
5EJJ
 
 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
5JI7
 
 | |
5JI9
 
 | |
5JIU
 
 | |
5JIA
 
 | |
4RCA
 
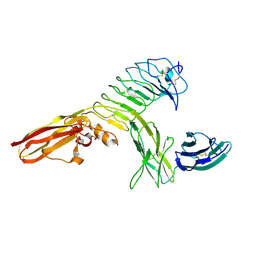 | | Crystal structure of human PTPdelta and human Slitrk1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 1, ... | | Authors: | Kim, H.M, Park, B.S, Kim, D, Lee, S.G. | | Deposit date: | 2014-09-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9908 Å) | | Cite: | Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion.
Nat Commun, 5, 2014
|
|
4RCW
 
 | |
8ITN
 
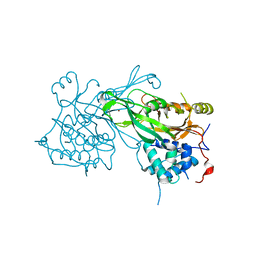 | | Crystal structure of USP47apo catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 47, ZINC ION | | Authors: | Kim, E.E, Shin, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of USP47 reveals a hot spot for inhibitor design.
Commun Biol, 6, 2023
|
|
8ITP
 
 | |
