3MYI
 
 | |
2IBF
 
 | |
1YDI
 
 | |
7KTU
 
 | |
7KTV
 
 | |
7KTT
 
 | |
7KTW
 
 | |
7UUW
 
 | | Cryogenic electron microscopy 3D map of F-actin bound by the Actin Binding Domain of alpha-catenin ortholog, HMP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The nematode alpha-catenin ortholog, HMP1, has an extended alpha-helix when bound to actin filaments.
J.Biol.Chem., 299, 2022
|
|
7UTJ
 
 | | Cryogenic electron microscopy 3D map of F-actin bound by human dimeric alpha-catenin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
7UXF
 
 | | Cryogenic electron microscopy 3D map of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
4DJ9
 
 | | Human vinculin head domain Vh1 (residues 1-258) in complex with the talin vinculin binding site 50 (VBS50, residues 2078-2099) | | Descriptor: | Talin-1, Vinculin | | Authors: | Yogesha, S.D, Rangarajan, E.S, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of vinculin in complex with vinculin binding site 50 (VBS50), the integrin binding site 2 (IBS2) of talin.
Protein Sci., 21, 2012
|
|
8EV1
 
 | | Dual Modulators | | Descriptor: | (3aR,4S,9bS)-4-(4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3aS,4R,9bR)-4-(4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, Estrogen Receptor, ... | | Authors: | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | Deposit date: | 2022-10-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
8EV2
 
 | | Dual Modulators | | Descriptor: | (3aS,4R,9bR)-4-(2-chloro-4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Estrogen receptor, ... | | Authors: | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | Deposit date: | 2022-10-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
6CDS
 
 | | Human neurofibromin 2/merlin/schwannomin residues 1-339 in complex with PIP2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Merlin, ... | | Authors: | Chinthalapudi, K, Sharff, A.J, Bricogne, G, Izard, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2.
Nat Commun, 9, 2018
|
|
4LQ8
 
 | |
4PR9
 
 | | Human Vinculin (residues 891-1066) in complex with PIP | | Descriptor: | GLYCEROL, Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Rangarajan, E.S, Patil, D, Izard, T. | | Deposit date: | 2014-03-05 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism and function of lipid-directed oligomerization of vinculin
To be Published
|
|
3H2V
 
 | | Human raver1 RRM1 domain in complex with human vinculin tail domain Vt | | Descriptor: | Raver-1, Vinculin | | Authors: | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | Deposit date: | 2009-04-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
3H2U
 
 | | Human raver1 RRM1, RRM2, and RRM3 domains in complex with human vinculin tail domain Vt | | Descriptor: | Raver-1, Vinculin | | Authors: | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | Deposit date: | 2009-04-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
6N9G
 
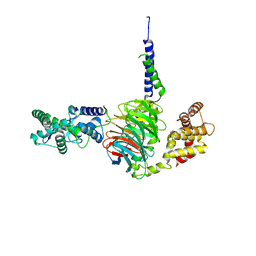 | | Crystal Structure of RGS7-Gbeta5 dimer | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Regulator of G-protein signaling 7 | | Authors: | Patil, D.N, Rangarajan, E, Izard, T, Martemyanov, K.A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Elife, 7, 2018
|
|
3LGE
 
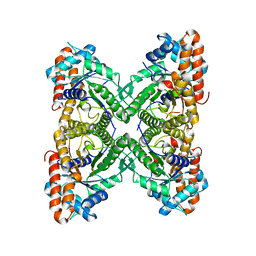 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
3RF3
 
 | |
2OT0
 
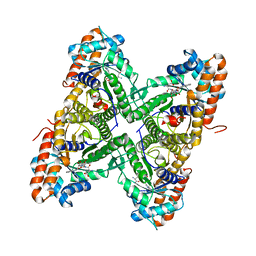 | |
2OT1
 
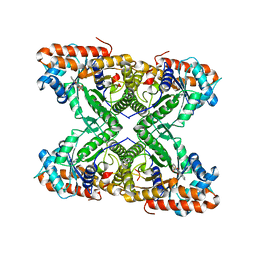 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with naphthol AS-E phosphate, a competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, N-(4-CHLOROPHENYL)-3-(PHOSPHONOOXY)NAPHTHALENE-2-CARBOXAMIDE | | Authors: | St-Jean, M, Izard, T, Sygusch, J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A hydrophobic pocket in the active site of glycolytic aldolase mediates interactions with wiskott-Aldrich syndrome protein.
J.Biol.Chem., 282, 2007
|
|
2PHL
 
 | | THE STRUCTURE OF PHASEOLIN AT 2.2 ANGSTROMS RESOLUTION: IMPLICATIONS FOR A COMMON VICILIN(SLASH)LEGUMIN STRUCTURE AND THE GENETIC ENGINEERING OF SEED STORAGE PROTEINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHASEOLIN, PHOSPHATE ION | | Authors: | Lawrence, M.C, Izard, T, Beuchat, M, Blagrove, R.J, Colman, P.M. | | Deposit date: | 1994-07-07 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of phaseolin at 2.2 A resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins.
J.Mol.Biol., 238, 1994
|
|
1TFU
 
 | |
