6YAJ
 
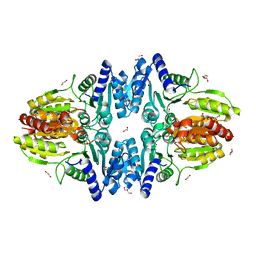 | | Split gene transketolase, inactive beta4 tetramer | | 分子名称: | 1,2-ETHANEDIOL, C-terminal chain of split transketolase from Carboxydothermus hydrogenoformans, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Isupov, M.N, Littlechild, J.A, James, P. | | 登録日 | 2020-03-12 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | 登録日 | 2016-12-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | 分子名称: | PEROXIREDOXIN-2 | | 著者 | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | 登録日 | 1999-10-07 | | 公開日 | 2000-07-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
8P2M
 
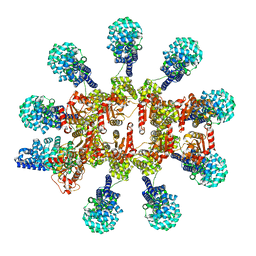 | | C. elegans TIR-1 protein. | | 分子名称: | NAD(+) hydrolase tir-1 | | 著者 | Isupov, M.N, Opatowsky, Y. | | 登録日 | 2023-05-16 | | 公開日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
8P2L
 
 | |
6T92
 
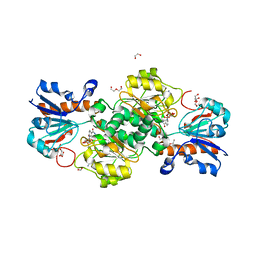 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | 登録日 | 2019-10-25 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
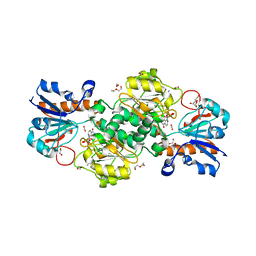 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | 登録日 | 2019-10-25 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T94
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH. | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | 登録日 | 2019-10-25 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | 登録日 | 2019-10-25 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
4BQ0
 
 | | Pseudomonas aeruginosa beta-alanine:pyruvate aminotransferase holoenzyme without divalent cations on dimer-dimer interface | | 分子名称: | BETA-ALANINE--PYRUVATE TRANSAMINASE, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Isupov, M.N, Lebedev, A.A, Westlake, A, Sayer, C, Littlechild, J.A. | | 登録日 | 2013-05-29 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Space-Group and Origin Ambiguity in Macromolecular Structures with Pseudo-Symmetry and its Treatment with the Program Zanuda.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7PNB
 
 | | Sulfolobus acidocaldarius 0406 filament. | | 分子名称: | 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sulfolobus acidocaldarius 0406 filament., beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Isupov, M.N, Gaines, M, Daum, B. | | 登録日 | 2021-09-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Electron cryo-microscopy reveals the structure of the archaeal thread filament.
Nat Commun, 13, 2022
|
|
8P60
 
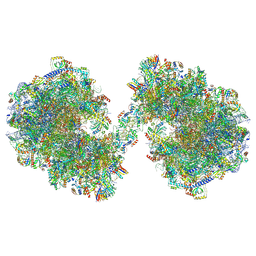 | | Spraguea lophii ribosome dimer | | 分子名称: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | 著者 | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | 登録日 | 2023-05-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (14.3 Å) | | 主引用文献 | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
8P5D
 
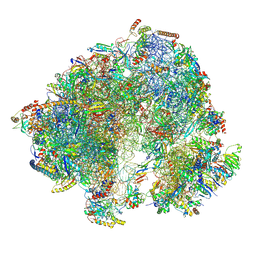 | | Spraguea lophii ribosome in the closed conformation by cryo sub tomogram averaging | | 分子名称: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | 著者 | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | 登録日 | 2023-05-23 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
4USM
 
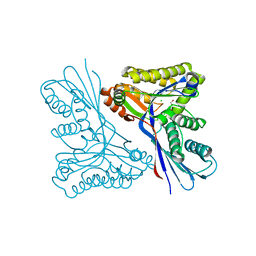 | | WcbL complex with glycerol bound to sugar site | | 分子名称: | CHLORIDE ION, GLYCEROL, PUTATIVE SUGAR KINASE | | 著者 | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | 登録日 | 2014-07-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
1H2B
 
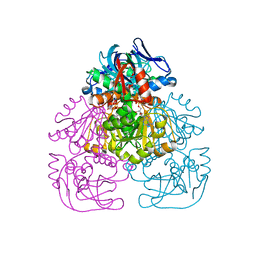 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | 分子名称: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | 著者 | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | 登録日 | 2002-08-02 | | 公開日 | 2003-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
6KXT
 
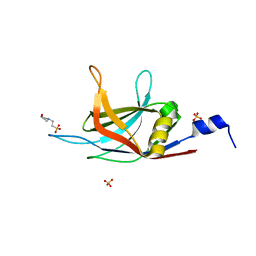 | | BON1-C2B | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein BONZAI 1, SULFATE ION | | 著者 | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | 登録日 | 2019-09-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXU
 
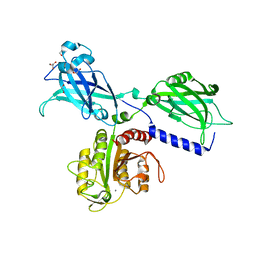 | | BON1 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | 登録日 | 2019-09-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXK
 
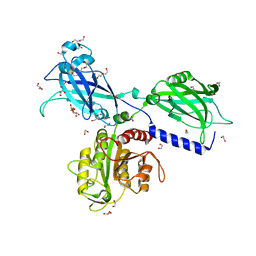 | | BON1 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | 登録日 | 2019-09-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6ZG0
 
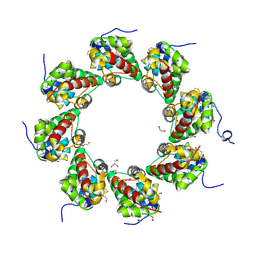 | | SARM1 SAM1-2 domains | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | 登録日 | 2020-06-18 | | 公開日 | 2020-11-11 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | 登録日 | 2020-06-18 | | 公開日 | 2020-11-11 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | 分子名称: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | 著者 | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | 登録日 | 2020-06-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
8EDD
 
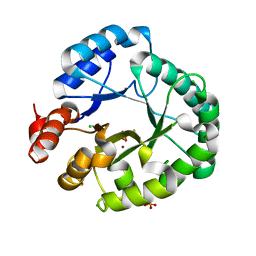 | | Staphylococcus aureus endonuclease IV Y33F mutant | | 分子名称: | CHLORIDE ION, FE (III) ION, PHOSPHATE ION, ... | | 著者 | Saper, M.A, Kirillov, S, Isupov, M.N, Wiener, R, Rouvinski, A. | | 登録日 | 2022-09-04 | | 公開日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
To Be Published
|
|
3LD0
 
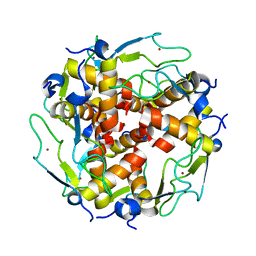 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | 分子名称: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | 著者 | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | 登録日 | 2010-01-12 | | 公開日 | 2010-02-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
5NFQ
 
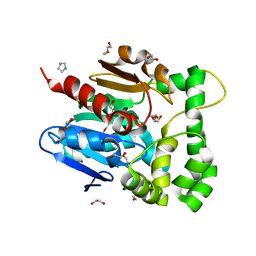 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | 著者 | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | 登録日 | 2017-03-15 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
7ANW
 
 | | hSARM1 NAD+ complex | | 分子名称: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | 登録日 | 2020-10-13 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
