5NPK
 
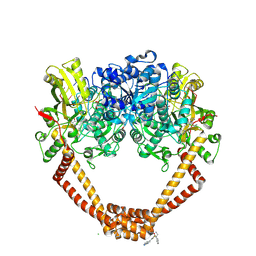 | | 1.98A STRUCTURE OF THIOPHENE1 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit B,DNA gyrase subunit B,DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-17 | | Release date: | 2017-07-12 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7MET
 
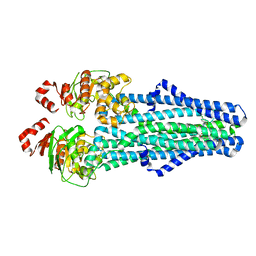 | | A. baumannii MsbA in complex with TBT1 decoupler | | Descriptor: | 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7MEW
 
 | | E. coli MsbA in complex with G247 | | Descriptor: | (2E)-3-{1-cyclopropyl-7-[(1S)-1-(3,6-dichloro-2-fluorophenyl)ethoxy]naphthalen-2-yl}prop-2-enoic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
5I8E
 
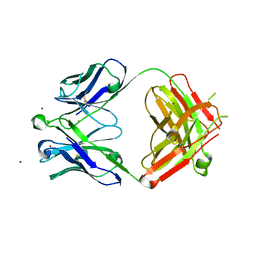 | | Crystal Structure of Broadly Neutralizing HIV-1 Fusion Peptide-Targeting Antibody VRC34.01 Fab | | Descriptor: | VRC34.01 Fab heavy chain, VRC34.01 Fab light chain, ZINC ION | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
5I8C
 
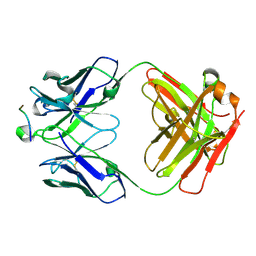 | | Crystal Structure of HIV-1 Clade A BG505 Fusion Peptide (residue 512-520) in Complex with Broadly Neutralizing Antibody VRC34.01 Fab | | Descriptor: | HIV-1 Clade A BG505 Fusion Peptide (residue 512-520), VRC34.01 Fab heavy chain, VRC34.01 Fab light chain | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
5I8H
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer in Complex with V3 Loop-targeting Antibody PGT122 Fab and Fusion Peptide-targeting Antibody VRC34.01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP.664 gp120, ... | | Authors: | Xu, K, Zhou, T, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
5IL0
 
 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL1
 
 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5JNY
 
 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
5JO5
 
 | |
5JR1
 
 | |
4MCD
 
 | | hinTrmD in complex with 5-PHENYLTHIENO[2,3-D]PYRIMIDIN-4(3H)-ONE | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lahiri, S. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
4MCC
 
 | | HinTrmD in complex with N-[4-(AMINOMETHYL)BENZYL]-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | N-[4-(aminomethyl)benzyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
4MCB
 
 | | H.influenzae TrmD in complex with N-(4-{[(1H-IMIDAZOL-2-YLMETHYL)AMINO]METHYL}BENZYL)-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | ACETATE ION, GLYCEROL, N-(4-{[(1H-imidazol-2-ylmethyl)amino]methyl}benzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, ... | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
5V0Y
 
 | | Solution structure of arenicin-3. | | Descriptor: | arenicin-3 | | Authors: | Edwards, I.A, Mobli, M. | | Deposit date: | 2017-02-28 | | Release date: | 2018-08-08 | | Last modified: | 2020-06-10 | | Method: | SOLUTION NMR | | Cite: | An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria
Nat Commun, 2020
|
|
6UBI
 
 | |
4Q6I
 
 | | Crystal structure of murine 2D5 Fab, a potent anti-CD4 HIV-1-neutralizing antibody in complex with CD4 | | Descriptor: | Heavy chain of murine 2D5 Fab, Light chain of murine 2D5 Fab, T-cell surface glycoprotein CD4 | | Authors: | Boyington, J.C, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor.
Sci Transl Med, 6, 2014
|
|
6UCH
 
 | | SMARCB1 nucleosome-interacting C-terminal alpha helix | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Valencia, A.M, Sun, Z.Y.J, Seo, H.S, Vangos, H.S, Yeoh, Z.C, Mashtalir, N, Dhe-Paganon, S, Kadoch, C. | | Deposit date: | 2019-09-16 | | Release date: | 2019-11-27 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling.
Cell, 179, 2019
|
|
6UCE
 
 | |
6UCF
 
 | |
4RQU
 
 | | Alcohol Dehydrogenase crystal structure in complex with NAD | | Descriptor: | Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
4RQT
 
 | | Alcohol Dehydrogenase Crystal Structure | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase class-P, SULFATE ION, ... | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
7RIT
 
 | | Drug-free A. baumannii MsbA | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-07-20 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
8PHI
 
 | | Crystal structure of prefusion-stabilized RSV F Variant DS-Cav1 in complex with Lonafarnib | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Rajak, M, Krey, T. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor.
Nat Commun, 15, 2024
|
|
