2D3G
 
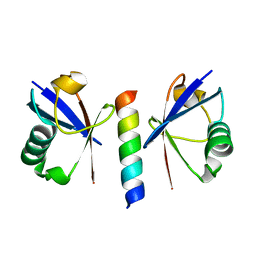 | | Double sided ubiquitin binding of Hrs-UIM | | 分子名称: | ubiquitin, ubiquitin interacting motif from hepatocyte growth factor-regulated tyrosine kinase substrate | | 著者 | Hirano, S, Kawasaki, M, Kato, R, Wakatsuki, S. | | 登録日 | 2005-09-28 | | 公開日 | 2005-12-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DX5
 
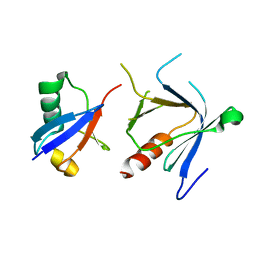 | | The complex structure between the mouse EAP45-GLUE domain and ubiquitin | | 分子名称: | Ubiquitin, Vacuolar protein sorting protein 36 | | 著者 | Hirano, S, Suzuki, N, Slagsvold, T, Kawasaki, M, Trambaiolo, D, Kato, R, Stenmark, H, Wakatsuki, S. | | 登録日 | 2006-08-24 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ZF3
 
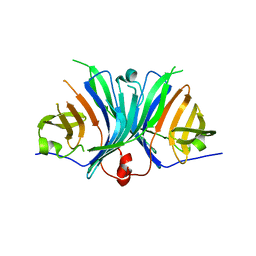 | | Crystal Structure of VioE | | 分子名称: | Hypothetical protein VioE | | 著者 | Hirano, S, Shiro, Y, Nagano, S. | | 登録日 | 2007-12-20 | | 公開日 | 2008-01-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of VioE, a key player in the construction of the molecular skeleton of violacein
To be Published
|
|
2ZF4
 
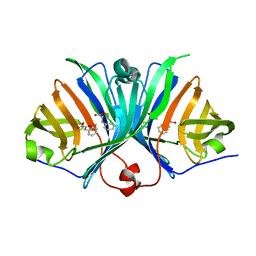 | |
5B2T
 
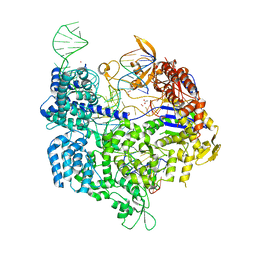 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2S
 
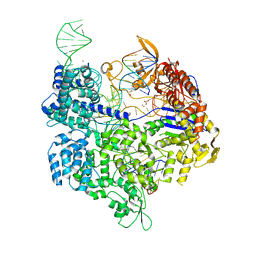 | | Crystal structure of the Streptococcus pyogenes Cas9 EQR variant in complex with sgRNA and target DNA (TGAG PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2R
 
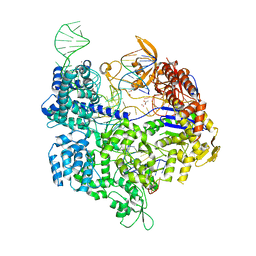 | | Crystal structure of the Streptococcus pyogenes Cas9 VQR variant in complex with sgRNA and target DNA (TGA PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
6JOO
 
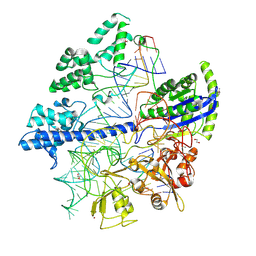 | | Crystal structure of Corynebacterium diphtheriae Cas9 in complex with sgRNA and target DNA | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated protein,CRISPR-associated endonuclease Cas9, Guide RNA, ... | | 著者 | Hirano, S, Ishitani, R, Nishimasu, H, Nureki, O. | | 登録日 | 2019-03-22 | | 公開日 | 2019-04-17 | | 最終更新日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9.
Nat Commun, 10, 2019
|
|
6IRZ
 
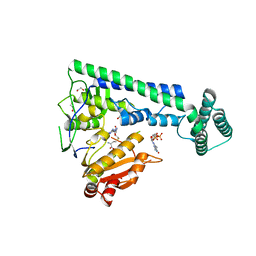 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | 分子名称: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-15 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRV
 
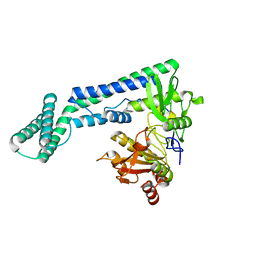 | |
6IS0
 
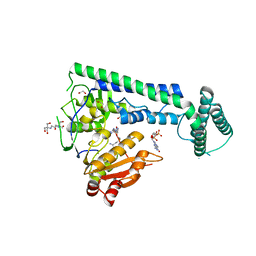 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | 分子名称: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-15 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRW
 
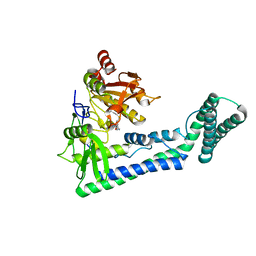 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRX
 
 | |
6IRY
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6AI6
 
 | | Crystal structure of SpCas9-NG | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | 著者 | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | 登録日 | 2018-08-21 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
6AEK
 
 | | Crystal structure of ENPP1 in complex with pApG | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-MONOPHOSPHATE, ... | | 著者 | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-08-05 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
6AEL
 
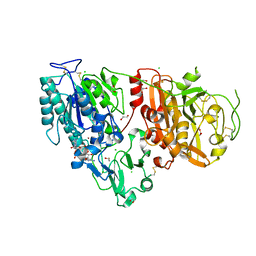 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | 著者 | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-08-05 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
6KR6
 
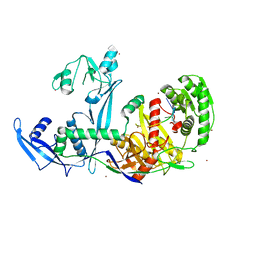 | | Crystal structure of Drosophila Piwi | | 分子名称: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | 著者 | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | 登録日 | 2019-08-21 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
8DMB
 
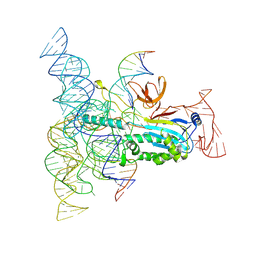 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | 分子名称: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | 著者 | Seiichi, H, Kappel, K, Zhang, F. | | 登録日 | 2022-07-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
2DWX
 
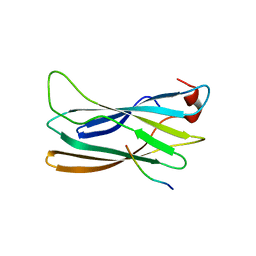 | | Co-crystal Structure Analysis of GGA1-GAE with the WNSF motif | | 分子名称: | ADP-ribosylation factor-binding protein GGA1, hinge peptide from ADP-ribosylation factor binding protein GGA1 | | 著者 | Inoue, M, Shiba, T, Yamada, Y, Ihara, K, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | 登録日 | 2006-08-21 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Molecular Basis for Autoregulatory Interaction Between GAE Domain and Hinge Region of GGA1
Traffic, 8, 2007
|
|
2DWY
 
 | | Crystal Structure Analysis of GGA1-GAE | | 分子名称: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA1 | | 著者 | Inoue, M, Shiba, T, Yamada, Y, Ihara, K, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | 登録日 | 2006-08-21 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular Basis for Autoregulatory Interaction Between GAE Domain and Hinge Region of GGA1
Traffic, 8, 2007
|
|
4XJ6
 
 | | Crystal structure of Escherichia coli DncV 3'-deoxy GTP bound form | | 分子名称: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, VC0179-like protein | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ3
 
 | | Crystal structure of Vibrio cholerae DncV GTP bound form | | 分子名称: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ1
 
 | | Crystal structure of Vibrio cholerae DncV apo form | | 分子名称: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ4
 
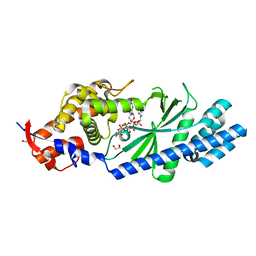 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy ATP bound form | | 分子名称: | 1,2-ETHANEDIOL, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | 著者 | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | 登録日 | 2015-01-08 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
