8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
5HGU
 
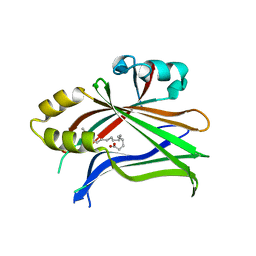 | |
1UEF
 
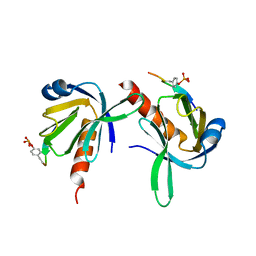 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Zhang, X, Yu, D, Wang, T. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
7Z8O
 
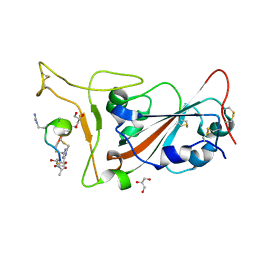 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8AAA
 
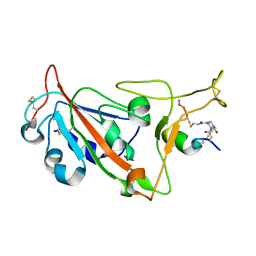 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6CDY
 
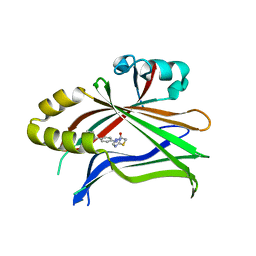 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
2R7B
 
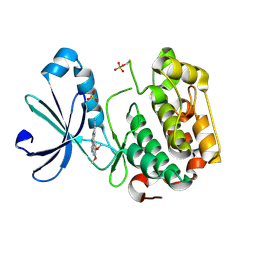 | | Crystal Structure of the Phosphoinositide-dependent Kinase-1 (PDK-1)Catalytic Domain bound to a dibenzonaphthyridine inhibitor | | Descriptor: | 10,11-dimethoxy-4-methyldibenzo[c,f]-2,7-naphthyridine-3,6-diamine, 3-phosphoinositide-dependent protein kinase 1, SULFATE ION | | Authors: | Olland, A.M. | | Deposit date: | 2007-09-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Dibenzo[c,f][2,7]naphthyridines as Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 Inhibitors.
J.Med.Chem., 50, 2007
|
|
8KHU
 
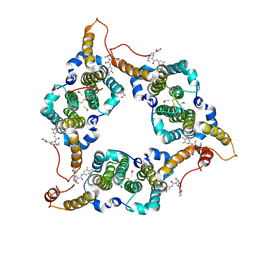 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
6VN2
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 18 | | Descriptor: | 1-({7-[(2R)-5-chloro-2-(piperazine-1-carbonyl)-2,3-dihydro-1-benzofuran-7-yl]thieno[3,2-b]pyridin-2-yl}methyl)-1H-pyrrole-2,5-dione, ACETATE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6VN3
 
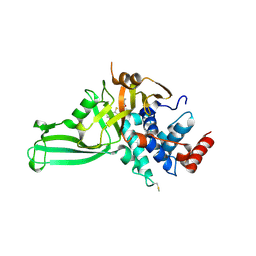 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 23 | | Descriptor: | 1-{[7-(5-chloro-2-{[(3R,4S)-4-fluoropyrrolidin-3-yl]oxy}-3-methylphenyl)thieno[3,2-b]pyridin-2-yl]methyl}-1H-pyrrole-2,5-dione, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6VN6
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 14 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION, [(2R)-5-chloro-7-{2-[(2S)-1-chloro-2,3-dihydroxypropan-2-yl]thieno[3,2-b]pyridin-7-yl}-2,3-dihydro-1-benzofuran-2-yl](piperazin-1-yl)methanone | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6VN5
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 7 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, [(2R)-7-(2-aminopyridin-4-yl)-5-chloro-2,3-dihydro-1-benzofuran-2-yl](piperazin-1-yl)methanone | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6VN4
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 1 | | Descriptor: | 3-({4-hydroxy-1-[(2R)-2-methyl-3-phenylpropanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6TI2
 
 | |
5M02
 
 | |
6GB5
 
 | | Structure of H-2Db with truncated SEV peptide and GL | | Descriptor: | Beta-2-microglobulin, GLY-LEU, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GB7
 
 | | Structure of H-2Db with scoop loop from tapasin | | Descriptor: | Beta-2-microglobulin, GLY-GLY-LEU-SER, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
