6DE6
 
 | |
4UV3
 
 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4UV2
 
 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
6FMD
 
 | | Targeting myeloid differentiation using potent human dihydroorotate dehydrogenase (hDHODH) inhibitors based on 2-hydroxypyrazolo[1,5-a]pyridine scaffold | | Descriptor: | 2-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Jarva, M, Andersson, M, Lolli, M.L, Friemann, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting Myeloid Differentiation Using Potent 2-Hydroxypyrazolo[1,5- a]pyridine Scaffold-Based Human Dihydroorotate Dehydrogenase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6F2U
 
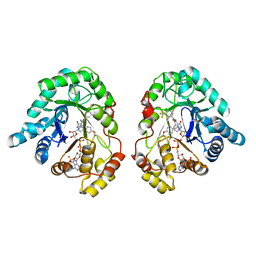 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 3-[(4-methoxyphenyl)methyl]-5-oxidanyl-~{N}-[3-(trifluoromethyl)phenyl]-1,2,3-triazole-4-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-11-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
6F78
 
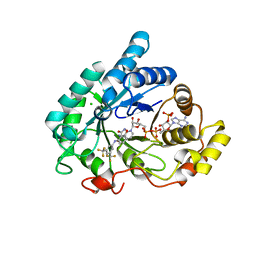 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: Application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 4-[[3,5-bis(trifluoromethyl)phenyl]amino]-1,2-benzoxazol-3-one, Aldo-keto reductase family 1 member C3, CHLORIDE ION, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
6GXK
 
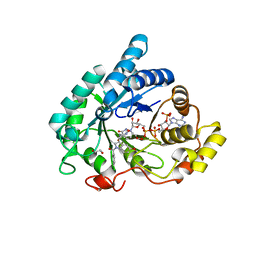 | | Crystal structure of Aldo-Keto Reductase 1C3 (AKR1C3) complexed with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 4-[[1-(4-chlorophenyl)carbonyl-5-methoxy-2-methyl-indol-3-yl]methyl]-1,2,5-oxadiazol-3-one, Aldo-keto reductase family 1 member C3, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bioisosteres of Indomethacin as Inhibitors of Aldo-Keto Reductase 1C3.
Acs Med.Chem.Lett., 10, 2019
|
|
5MVD
 
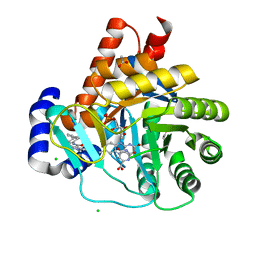 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 1,5-dimethyl-3-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
5MVC
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 4-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,5-thiadiazole-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
5MUT
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
6I55
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) co-crystallized with N-(2,2-Diphenylethyl)-4-hydroxy-1,2,5-thiadiazole-3-carboxamide | | Descriptor: | Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, N-(2,2-Diphenylethyl)-4-hydroxy-1,2,5-thiadiazole-3-carboxamide, ... | | Authors: | Goyal, P, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Hydroxyazole scaffold-based Plasmodium falciparum dihydroorotate dehydrogenase inhibitors: Synthesis, biological evaluation and X-ray structural studies.
Eur J Med Chem, 163, 2018
|
|
6I4B
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) co-crystallized with 3-Hydroxy-1-methyl-5-((3-(trifluoromethyl)phenoxy)methyl)-1H-pyrazole-4-carboxylic acid | | Descriptor: | 1-methyl-3-oxidanyl-5-[[3-(trifluoromethyl)phenoxy]methyl]pyrazole-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Goyal, P, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S. | | Deposit date: | 2018-11-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Hydroxyazole scaffold-based Plasmodium falciparum dihydroorotate dehydrogenase inhibitors: Synthesis, biological evaluation and X-ray structural studies.
Eur J Med Chem, 163, 2018
|
|
4O7K
 
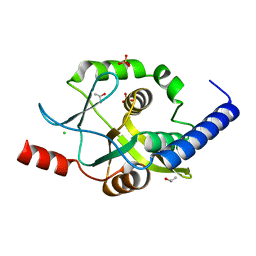 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, PHOSPHATE ION, ... | | Authors: | Maindola, P, Goyal, P, Arulandu, A. | | Deposit date: | 2013-12-25 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
4OVB
 
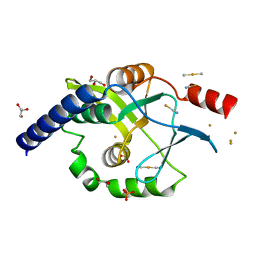 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor, Gold (I) Cyanide derivative | | Descriptor: | GLYCEROL, GOLD (I) CYANIDE ION, PHOSPHATE ION, ... | | Authors: | Maindola, P, Goyal, P, Arulandu, A. | | Deposit date: | 2014-02-21 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
7EPI
 
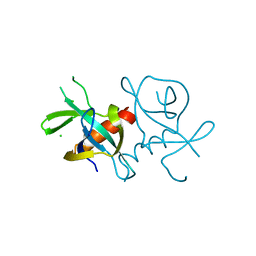 | |
7EPJ
 
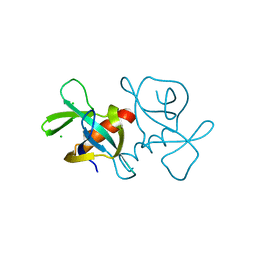 | |
5NV9
 
 | | Substrate-bound outward-open state of a Na+-coupled sialic acid symporter reveals a novel Na+-site | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, N-acetyl-beta-neuraminic acid, PHOSPHATE ION, ... | | Authors: | Wahlgren, W.Y, North, R.A, Dunevall, E, Paz, A, Goyal, P, Bisignano, P, Grabe, M, Dobson, R, Abramson, J, Ramaswamy, S, Friemann, R. | | Deposit date: | 2017-05-03 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+site.
Nat Commun, 9, 2018
|
|
5NVA
 
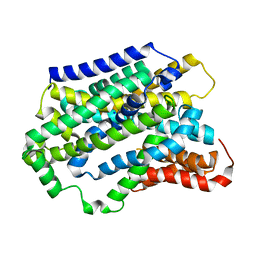 | | Substrate-bound outward-open state of a Na+-coupled sialic acid symporter reveals a novel Na+-site | | Descriptor: | N-acetyl-beta-neuraminic acid, Putative sodium:solute symporter, SODIUM ION | | Authors: | Wahlgren, W.Y, North, R.A, Dunevall, E, Goyal, P, Grabe, M, Dobson, R, Abramson, J, Ramaswamy, S, Friemann, R. | | Deposit date: | 2017-05-03 | | Release date: | 2018-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+site.
Nat Commun, 9, 2018
|
|
6H4E
 
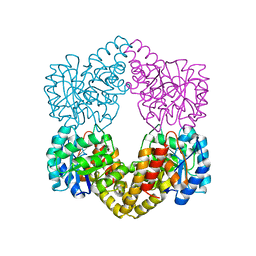 | | Proteus mirabilis N-acetylneuraminate lyase | | Descriptor: | Putative N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Garcia-Bonete, M.J, Goyal, P, Katona, G, Dobson, R.C.J, Friemann, R. | | Deposit date: | 2018-07-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | The structure of Proteus mirabilis N-acetylneuraminate lyase reveals an intermolecular disulphide bond
To Be Published
|
|
6TZW
 
 | |
6OFP
 
 | |
8H73
 
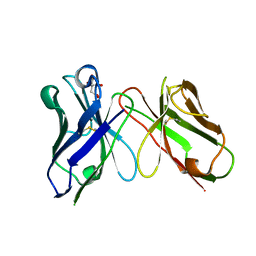 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | CITRATE ANION, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3C
 
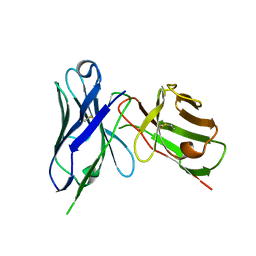 | | Crystal structure of M2e Influenza peptide in complex with antibody scFv | | Descriptor: | Matrix protein 2, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3B
 
 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | GLYCEROL, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
7EPG
 
 | |
